6XT1
 
 | |
6ORH
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
2OZN
 
 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1OIF
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, BETA-GLUCOSIDASE | | Authors: | Gloster, T, Zechel, D, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-16 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1OIM
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 1-DEOXYNOJIRIMYCIN, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1OIN
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1OD0
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
2O4E
 
 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
6BIA
 
 | | Crystal structure of Ps i-CgsB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B1V
 
 | | Crystal structure of Ps i-CgsB C78S in complex with i-neocarratetraose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B0J
 
 | | Crystal structure of Ps i-CgsB in complex with k-i-k-neocarrahexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B0K
 
 | | Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6DMS
 
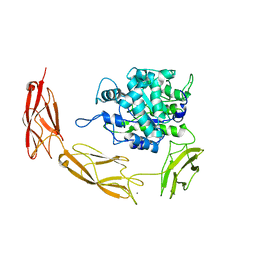 | | Endo-fucoidan hydrolase MfFcnA4_H294Q from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
8U5O
 
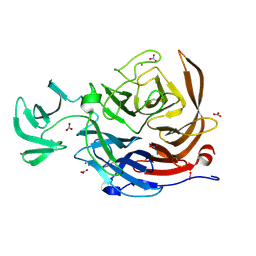 | | The structure of the catalytic domain of NanI sialdase in complex with Neu5Gc | | Descriptor: | CALCIUM ION, Exo-alpha-sialidase, N-glycolyl-alpha-neuraminic acid, ... | | Authors: | Medley, B.J, Low, K.E, Garber, J.M, Gray, T.E, Liu, L, Klassen, L, Fordwour, O.B, Inglis, G.D, Boons, G.J, Zandberg, W.F, Abbott, W.D, Boraston, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A terminal case of glycan catabolism structural and enzymatic characterization of the
sialidases of Clostridium perfringens
To Be Published
|
|
8U2A
 
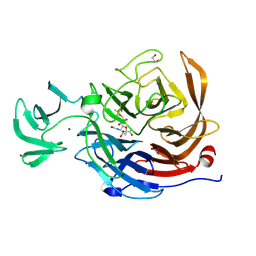 | | Crystal structure of NanI in complex with Neu5,9Ac | | Descriptor: | 1,2-ETHANEDIOL, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, Exo-alpha-sialidase, ... | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A terminal case of glycan catabolism:
structural and enzymatic characterization of the
sialidases of Clostridium perfringens
To Be Published
|
|
1J84
 
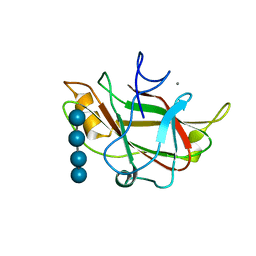 | | STRUCTURE OF FAM17 CARBOHYDRATE BINDING MODULE FROM CLOSTRIDIUM CELLULOVORANS WITH BOUND CELLOTETRAOSE | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta glucanase EngF | | Authors: | Notenboom, V, Boraston, A.B, Chiu, P, Freelove, A.C.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-05-20 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study.
J.Mol.Biol., 314, 2001
|
|
1KNM
 
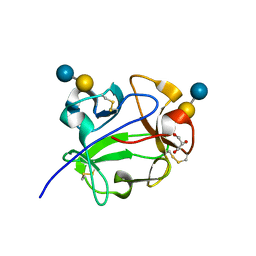 | | Streptomyces lividans Xylan Binding Domain cbm13 in Complex with Lactose | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KNL
 
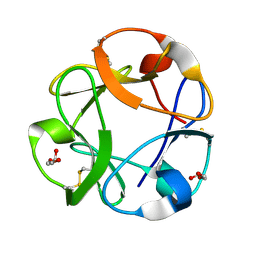 | | Streptomyces lividans Xylan Binding Domain cbm13 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1J83
 
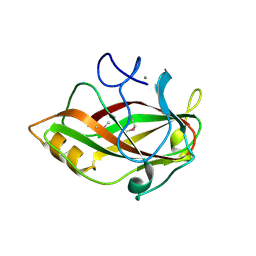 | | STRUCTURE OF FAM17 CARBOHYDRATE BINDING MODULE FROM CLOSTRIDIUM CELLULOVORANS | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA GLUCANASE ENGF | | Authors: | Notenboom, V, Boraston, A.B, Chiu, P, Freelove, A.C.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-05-20 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study.
J.Mol.Biol., 314, 2001
|
|
1MC9
 
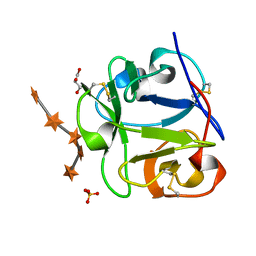 | | STREPROMYCES LIVIDANS XYLAN BINDING DOMAIN CBM13 IN COMPLEX WITH XYLOPENTAOSE | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, SULFATE ION, ... | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2002-08-06 | | Release date: | 2002-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
5FQE
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
9C20
 
 | | The Sialidase NanJ in complex with Neu5,9Ac | | Descriptor: | 1,2-ETHANEDIOL, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, exo-alpha-sialidase | | Authors: | Medley, B.J, Low, K.E, Garber, J.M, Gray, T.E, Leeann, L.L, Fordwour, O.B, Inglis, G.D, Boons, G.J, Zandberg, W.F, Abbott, W, Boraston, A. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | NanJ sialidase in complex with Neu5,9Ac
To Be Published
|
|
3REN
 
 | | CPF_2247, a novel alpha-amylase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase, family 8, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Boraston, A.B. | | Deposit date: | 2011-04-04 | | Release date: | 2011-05-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of CPF_2247, a novel alpha-amylase from Clostridium perfringens.
Proteins, 79, 2011
|
|
3QSP
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 non-productive substrate complex with alpha-1,6-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
5FR0
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
