1IMT
 
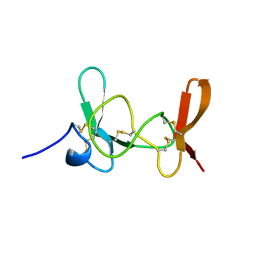 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
8B7I
 
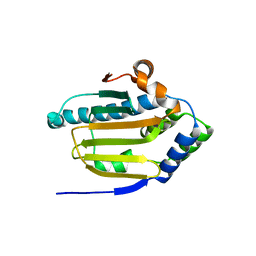 | | Human HSP90 alpha ATP Binding Domain, ATP-lid open conformation, R60A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brutscher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
8B7J
 
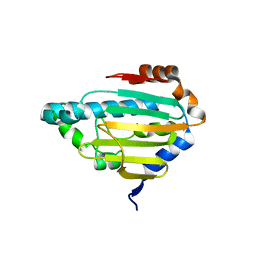 | | Human HSP90 alpha ATP Binding Domain, ATP-lid closed conformation, R46A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brustcher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
6F3K
 
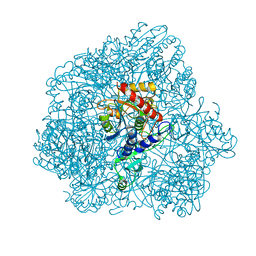 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
2RN1
 
 | | Liquid crystal solution structure of the kissing complex formed by the apical loop of the HIV TAR RNA and a high affinity RNA aptamer optimized by SELEX | | Descriptor: | RNA (5'-R(P*GP*AP*GP*CP*CP*CP*UP*GP*GP*GP*AP*GP*GP*CP*UP*C)-3'), RNA (5'-R(P*GP*CP*UP*GP*GP*UP*CP*CP*CP*AP*GP*AP*CP*AP*GP*C)-3') | | Authors: | Van Melckebeke, H, Devany, M, Di Primo, C, Beaurain, F, Toulme, J, Bryce, D.L, Boisbouvier, J. | | Deposit date: | 2007-12-05 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1QKY
 
 | | Solution structure of PI7, a non toxic peptide isolated from the scorpion Pandinus Imperator. | | Descriptor: | TOXIN 7 FROM PANDINUS IMPERATOR | | Authors: | Delepierre, M, Prochnicka-Chalufour, A, Boisbouvier, J, Possani, L.D. | | Deposit date: | 1999-08-17 | | Release date: | 2000-02-03 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Pi7, an orphan peptide from the scorpion Pandinus imperator: a 1H-NMR analysis using a nano-NMR Probe.
Biochemistry, 38, 1999
|
|
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
2L2N
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for the first dsRBD of protein HYL1 | | Descriptor: | Hyponastic leave 1 | | Authors: | Rasia, R.M, Mateos, J.L, Bologna, N.G, Burdisso, P, Imbert, L, Palatnik, J.F, Boisbouvier, J. | | Deposit date: | 2010-08-23 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the Plant MicroRNA Processing-Associated Protein HYL1.
Biochemistry, 49, 2010
|
|
2L2M
 
 | | Solution structure of the second dsRBD of HYL1 | | Descriptor: | Hyponastic leave 1 | | Authors: | Rasia, R.M, Mateos, J.L, Bologna, N.G, Burdisso, P, Imbert, L, Palatnik, J.F, Boisbouvier, J. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the Plant MicroRNA Processing-Associated Protein HYL1.
Biochemistry, 49, 2010
|
|
2GBH
 
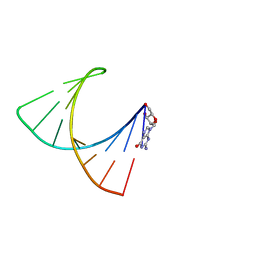 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
2LRS
 
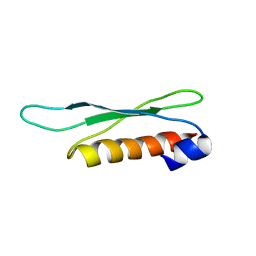 | | The second dsRBD domain from A. thaliana DICER-LIKE 1 | | Descriptor: | Endoribonuclease Dicer homolog 1 | | Authors: | Burdisso, P, Suarez, I, Bersch, B, Bologna, N, Palatnik, J, Boisbouvier, J, Rasia, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Second Double-Stranded RNA Binding Domain of Dicer-like Ribonuclease 1: Structural and Biochemical Characterization.
Biochemistry, 51, 2012
|
|
