7PG2
 
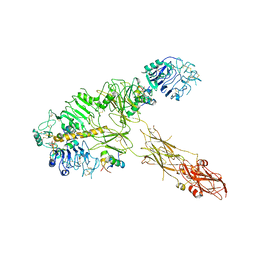 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG0
 
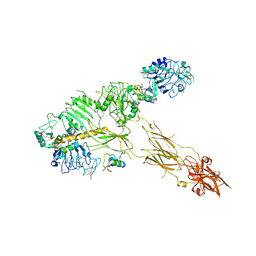 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin with visible ddm micelle, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG4
 
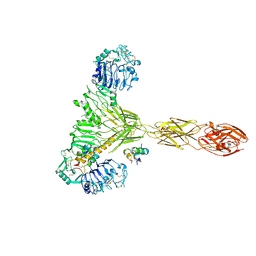 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 2 insulin, conf 3 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG3
 
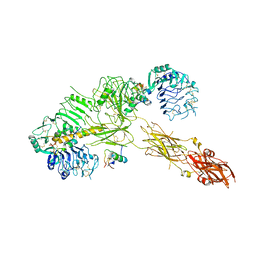 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 3 insulin, conf 2 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
4A1I
 
 | | ykud from B.subtilis | | Descriptor: | CADMIUM ION, CHLORIDE ION, PUTATIVE L, ... | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
7Z17
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
4D40
 
 | | High-Resolution Structure of a Type IV Pilin from Shewanella oneidensis | | Descriptor: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | Authors: | Gorgel, M, JensenUlstrup, J, Boeggild, A, Nissen, P, Boesen, T. | | Deposit date: | 2014-10-24 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
4A1J
 
 | | Ykud L,D-transpeptidase from B.subtilis | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Ykud
To be Published
|
|
4A1K
 
 | | Ykud L,D-transpeptidase | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
1ZM3
 
 | | Structure of the apo eEF2-ETA complex | | Descriptor: | Elongation factor 2, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
1ZM9
 
 | | Structure of eEF2-ETA in complex with PJ34 | | Descriptor: | Elongation factor 2, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
1ZM4
 
 | | Structure of the eEF2-ETA-bTAD complex | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, Elongation factor 2, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
1ZM2
 
 | | Structure of ADP-ribosylated eEF2 in complex with catalytic fragment of ETA | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
8A7D
 
 | | Partial dimer complex of PAPP-A and its inhibitor STC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Pappalysin-1, ... | | Authors: | Kobbero, S.D, Gajhede, M, Mirza, O.A, Boesen, T, Oxvig, C. | | Deposit date: | 2022-06-20 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism.
Nat Commun, 13, 2022
|
|
8A7E
 
 | | PAPP-A dimer in complex with its inhibitor STC2 | | Descriptor: | CALCIUM ION, Pappalysin-1, Stanniocalcin-2, ... | | Authors: | Kobbero, S.D, Gajhede, M, Mirza, O.A, Boesen, T, Oxvig, C. | | Deposit date: | 2022-06-20 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.02 Å) | | Cite: | Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism.
Nat Commun, 13, 2022
|
|
