1PFP
 
 | | CATHELIN-LIKE MOTIF OF PROTEGRIN-3 | | Descriptor: | Protegrin 3 | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
1PAE
 
 | | nucleoside diphosphate kinase | | Descriptor: | Nucleoside diphosphate kinase, cytosolic, SELENIUM ATOM | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
1DNG
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL ACIDIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DN3
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
2L3Z
 
 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
6RTB
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2B | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6SSX
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-09-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6SST
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2B | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-09-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6RT0
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
2RLZ
 
 | | Solid-State MAS NMR structure of the dimer Crh | | Descriptor: | HPr-like protein crh | | Authors: | Loquet, A, Bardiaux, B, Gardiennet, C, Blanchet, C, Baldus, M, Nilges, M, Malliavin, T, Bockmann, A. | | Deposit date: | 2007-09-04 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | 3D structure determination of the Crh protein from highly ambiguous solid-state NMR restraints.
J.Am.Chem.Soc., 130, 2008
|
|
2N1F
 
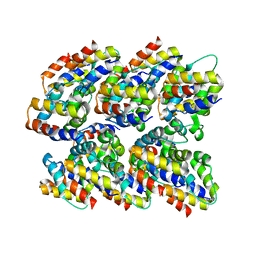 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1UXD
 
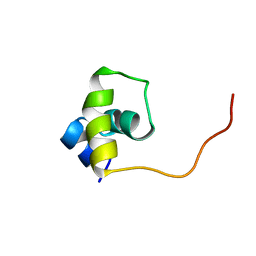 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1UXC
 
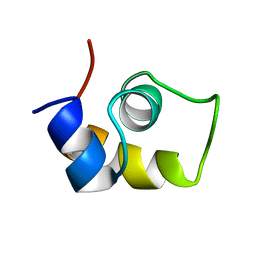 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
2LBU
 
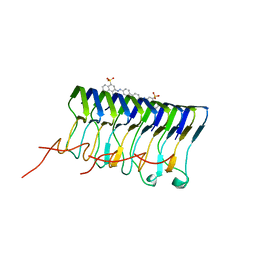 | | HADDOCK calculated model of Congo red bound to the HET-s amyloid | | Descriptor: | Small s protein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Schutz, A.K, Soragni, A, Hornemann, S, Aguzzi, A, Ernst, M, Bockmann, A, Meier, B.H. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-20 | | Method: | SOLID-STATE NMR | | Cite: | The Amyloid-Congo Red Interface at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 2011
|
|
2NAO
 
 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
