1Z0R
 
 | | Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transcription-State Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Bobay, B.G, Andreeva, A, Mueller, G.A, Cavanagh, J, Murzin, A.G. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
Febs Lett., 579, 2005
|
|
2JVK
 
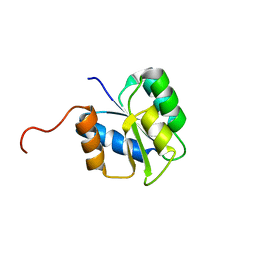 | |
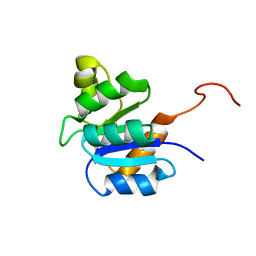
2JVJ
 
 | |
2JVI
 
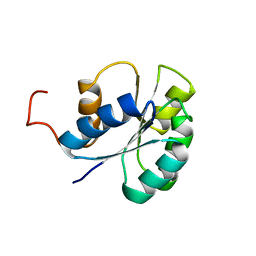 | |
2MIG
 
 | |
2LZF
 
 | |
2MFO
 
 | |
2MID
 
 | |
2MFM
 
 | |
2MIE
 
 | |
2MIH
 
 | |
2MIF
 
 | |
5TN2
 
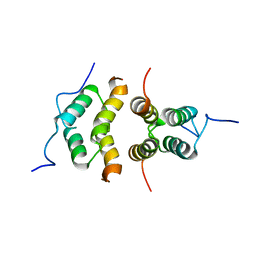 | | Solution Structure of the C-terminal multimerization domain of the master biofilm-regulator SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator SinR | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
5TN0
 
 | | Solution Structure of the N-terminal DNA-binding domain of the master biofilm-regulator SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator SinR | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
5TMX
 
 | | Solution Structure of SinI, antagonist to the master biofilm-regulator SinR in Bacillus subtilis | | Descriptor: | Protein SinI | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
2RO4
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO3
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator Abh | | Descriptor: | Putative transition state regulator abh | | Authors: | Sullivan, D.M, Bobay, B.G, Douglas, K.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO5
 
 | | RDC-refined solution structure of the N-terminal DNA recognition domain of the Bacillus subtilis transition-state regulator SpoVT | | Descriptor: | Stage V sporulation protein T | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2FY9
 
 | |
2K1N
 
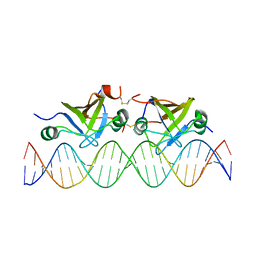 | | DNA bound structure of the N-terminal domain of AbrB | | Descriptor: | AbrB family transcriptional regulator, DNA (25-MER) | | Authors: | Cavanagh, J, Bobay, B.G, Sullivan, D.M, Thompson, R.J. | | Deposit date: | 2008-03-10 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Insights into the Nature of DNA Binding of AbrB-like Transcription Factors
Structure, 16, 2008
|
|
2KRF
 
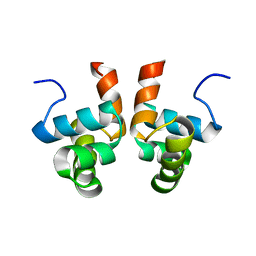 | | NMR solution structure of the DNA binding domain of Competence protein A | | Descriptor: | Transcriptional regulatory protein comA | | Authors: | Hobbs, C.A, Bobay, B.G, Thompson, R.J, Perego, M, Cavanagh, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and DNA-binding model of the DNA-binding domain of competence protein A.
J.Mol.Biol., 398, 2010
|
|
2MX1
 
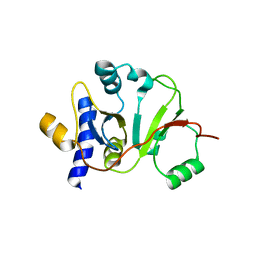 | | Structure of the E. coli Threonylcarbamoyl-AMP Synthase TSAC | | Descriptor: | Threonylcarbamoyl-AMP synthase | | Authors: | Harris, K.A, Bobay, B.G, Sarachan, K.L, Sims, A.F, Bilbille, Y, Deutsch, C, Iwata-Reuyl, D, Agris, P.F. | | Deposit date: | 2014-12-06 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-based Structural Analysis of Threonylcarbamoyl-AMP Synthase and Its Substrate Interactions.
J.Biol.Chem., 290, 2015
|
|
2NAZ
 
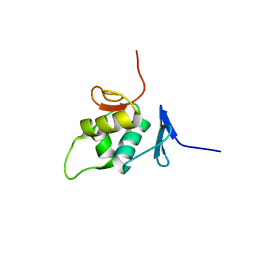 | | The solution NMR structure of the C-terminal effector domain of BfmR from Acinetobacter baumannii | | Descriptor: | Transcriptional regulatory protein RstA | | Authors: | Olson, A.L, Thompson, R.J, Cavanagh, J, Feldmann, E.A, Bobay, B.G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Biofilm-controlling Response Regulator BfmR from Acinetobacter baumannii Reveals Details of Its DNA-binding Mechanism.
J.Mol.Biol., 430, 2018
|
|
