6WQP
 
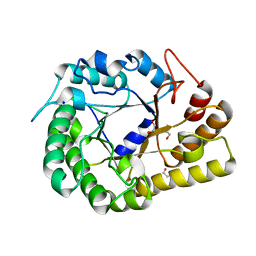 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Endoglucanase, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQV
 
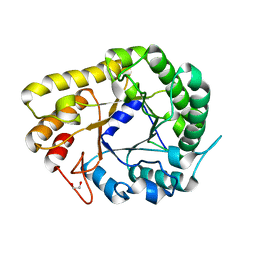 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
8SF6
 
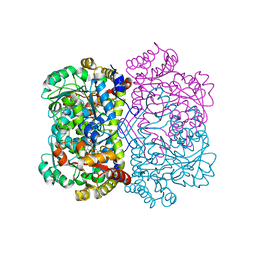 | | Promiscuous amino acid gamma synthase from Caldicellulosiruptor hydrothermalis in closed conformation | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Buller, A.R, Zmich, A.P, Bingman, C.A. | | Deposit date: | 2023-04-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Multiplexed Assessment of Promiscuous Non-Canonical Amino Acid Synthase Activity in a Pyridoxal Phosphate-Dependent Protein Family.
Acs Catalysis, 13, 2023
|
|
8SF5
 
 | |
6UI3
 
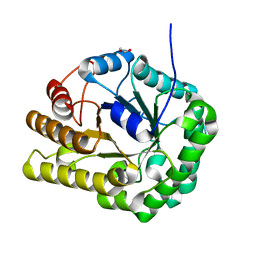 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6NIV
 
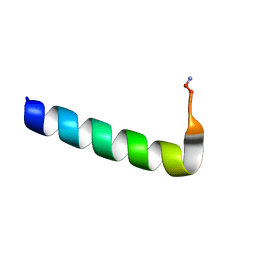 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5HOQ
 
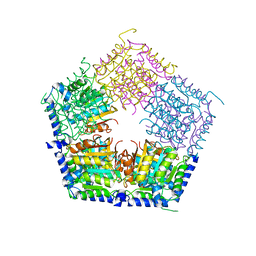 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
1Q4R
 
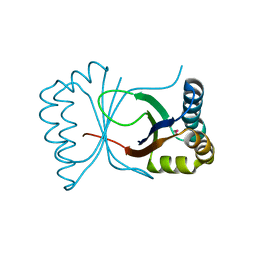 | | Gene Product of At3g17210 from Arabidopsis Thaliana | | Descriptor: | MAGNESIUM ION, protein At3g17210 | | Authors: | Phillips Jr, G.N, Bingman, C.A, Johnson, K.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana
Proteins, 57, 2004
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
5W4D
 
 | | C. japonica N-domain, Selenomethionine mutant | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CHLORIDE ION, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
5W4A
 
 | | C. japonica N-domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
5TD6
 
 | | C. elegans FOG-3 BTG/Tob domain - H47N, C117A | | Descriptor: | FOG-3 protein, SULFATE ION | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2016-09-17 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | An RNA-Binding Multimer Specifies Nematode Sperm Fate.
Cell Rep, 23, 2018
|
|
6OWD
 
 | |
4FE3
 
 | |
6PZ7
 
 | | GH5-4 broad specificity endoglucanase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Endoglucanase family 5 | | Authors: | Bianchetti, C.M, Bingman, C.A, Fox, B.G. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6Q1I
 
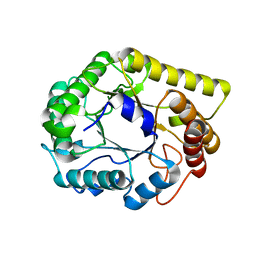 | |
6O4M
 
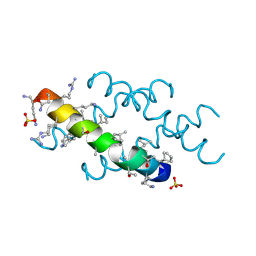 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
4G2T
 
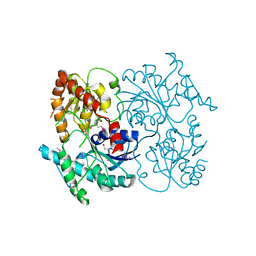 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | Descriptor: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4FZR
 
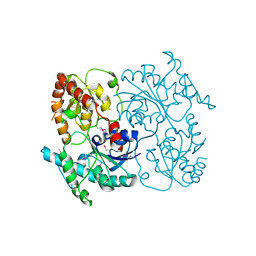 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4HPV
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
