7BX2
 
 | |
2K98
 
 | |
2K8O
 
 | |
5Z32
 
 | | LPS bound solution NMR structure of WS2-VR18 | | Descriptor: | VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY-LYS-ASN-LYS-SER-ARG | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
5Z31
 
 | | LPS bound solution structure of WS2-KG18 | | Descriptor: | LYS-ASN-LYS-SER-ARG-VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
6KH8
 
 | | Solution structure of Zn free Bovine Pancreatic Insulin in 20% acetic acid-d4 (pH 1.9) | | Descriptor: | Insulin A Chain, Insulin B chain | | Authors: | Bhunia, A, Ratha, B.N, Kar, R.K, Brender, J.R. | | Deposit date: | 2019-07-14 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
5ZYX
 
 | | Solution NMR structure of K30 peptide in 10 mM dioctanoyl phosphatidylglycerol (D8PG) | | Descriptor: | ARG-TRP-LYS-ARG-HIS-ILE-SER-GLU-GLN-LEU-ARG-ARG-ARG-ASP-ARG-LEU-GLN-ARG-GLN-ALA | | Authors: | Bhunia, A, Mohid, A, Stella, L, Calligari, P. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, Antibacterial Potential, and Structural Characterization of N-Acylated Derivatives of the Human Autophagy 16 Polypeptide.
Bioconjug.Chem., 30, 2019
|
|
2MWL
 
 | |
2NAU
 
 | |
2NAT
 
 | |
2MD3
 
 | |
2MD4
 
 | |
2MD2
 
 | |
2MD1
 
 | |
2MXG
 
 | |
2N63
 
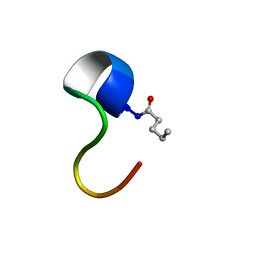 | | Structure of C4VG16KRKP | | Descriptor: | PENTANOIC ACID, antimicrobial peptide C4VG16KRKP | | Authors: | Bhunia, A, Datta, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding.
J Colloid Interface Sci, 461, 2016
|
|
2N9N
 
 | |
2KNS
 
 | |
2N9M
 
 | |
2MXH
 
 | |
2L36
 
 | |
2KGN
 
 | |
2LQ2
 
 | | Solution structure of de novo designed peptide 4m | | Descriptor: | de novo designed antifreeze peptide 4m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2L4U
 
 | |
2LQ1
 
 | | Solution structure of de novo designed antifreeze peptide 3 | | Descriptor: | de novo designed antifreeze peptide 3 | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
