4B55
 
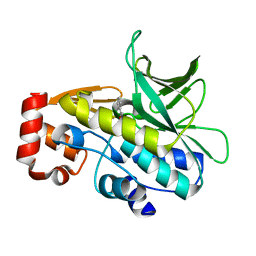 | | Crystal Structure of the Covalent Adduct Formed between Mycobacterium marinum Aryalamine N-acetyltransferase and Phenyl vinyl ketone a derivative of Piperidinols | | Descriptor: | 3-hydroxy-1-phenylpropan-1-one, ARYLAMINE N-ACETYLTRANSFERASE NAT | | Authors: | Abuhammad, A, Fullam, E, Lowe, E.D, Staunton, D, Kawamura, A, Westwood, I.M, Bhakta, S, Garner, A.C, Wilson, D.L, Seden, P.T, Davies, S.G, Russell, A.J, Garman, E.F, Sim, E. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Piperidinols that Show Anti-Tubercular Activity as Inhibitors of Arylamine N-Acetyltransferase: An Essential Enzyme for Mycobacterial Survival Inside Macrophages.
Plos One, 7, 2012
|
|
5JZS
 
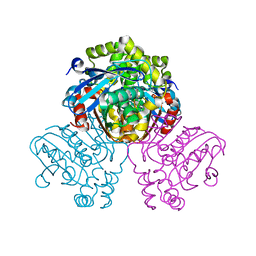 | | HsaD bound to 3,5-dichloro-4-hydroxybenzoic acid | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD, 3,5-dichloro-4-hydroxybenzoic acid | | Authors: | Ryan, A, Polycarpou, E, Lack, N, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E.D, Ballet, R, Abuhammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZB
 
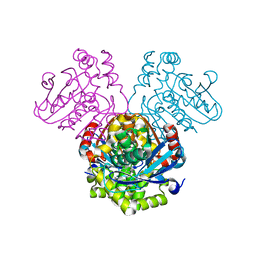 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZ9
 
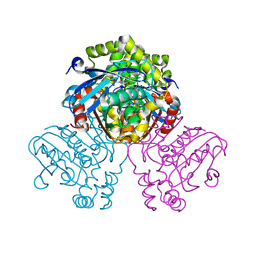 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
7D80
 
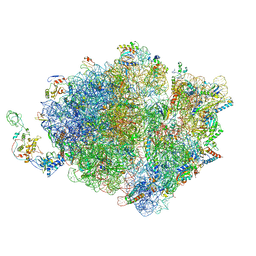 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
7D6Z
 
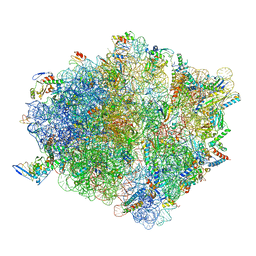 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase and trigger factor | | Descriptor: | 16S ribosomal rRNA, 23S ribosomal rRNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
2XJA
 
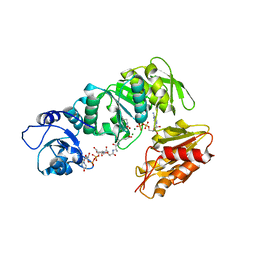 | | Structure of MurE from M.tuberculosis with dipeptide and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Basavannacharya, C, Moody, P.R, Bhakta, S, Keep, N. | | Deposit date: | 2010-07-03 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Essential Residues for the Enzyme Activity of ATP-Dependent Mure Ligase from Mycobacterium Tuberculosis.
Protein Cell, 1, 2010
|
|
4WJT
 
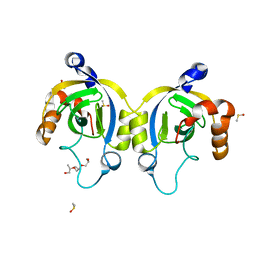 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG | | Descriptor: | (2S)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLK
 
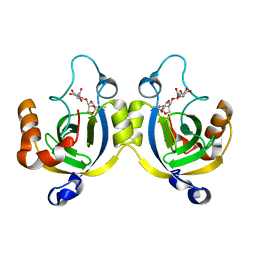 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with reaction product | | Descriptor: | N-[(1R,2S,3R,4R,5R)-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-4-yl]ethanamide, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLI
 
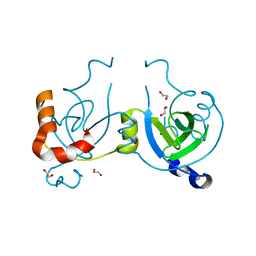 | | Stationary Phase Survival Protein YuiC from B.subtilis | | Descriptor: | 1,2-ETHANEDIOL, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
1W4T
 
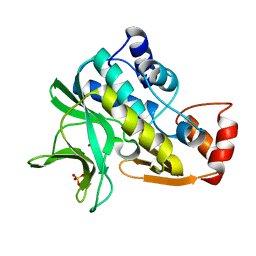 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
2XW7
 
 | | Structure of Mycobacterium smegmatis putative reductase MS0308 | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Evangelopoulos, D, Gupta, A, Lack, N, Cronin, N, Daviter, T, Sim, E, Keep, N.H, Bhakta, S. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of an Oxidoreductase from the Arylamine N-Acetyltransferase Operon in Mycobacterium Smegmatis.
FEBS J., 278, 2011
|
|
2WTZ
 
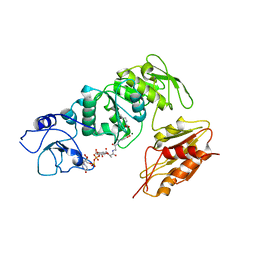 | | MurE ligase of Mycobacterium Tuberculosis | | Descriptor: | MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE-D-GLUTAMATE | | Authors: | Basavannacharya, C, Robertson, G, Munshi, T, Keep, N.H, Bhakta, S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP-Dependent Mure Ligase in Mycobacterium Tuberculosis: Biochemical and Structural Characterisation.
Tuberculosis(Edinb.), 90, 2010
|
|
6IY7
 
 | |
6J45
 
 | |
6IZ7
 
 | |
6IZI
 
 | |
6J0A
 
 | |
6JR3
 
 | |
1W5R
 
 | | X-ray crystallographic structure of a C70Q Mycobacterium smegmatis N- arylamine Acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Holton, S.J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E.M, Sim, E. | | Deposit date: | 2004-08-09 | | Release date: | 2005-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Investigation of the Catalytic Triad of Arylamine N-Acetyltransferases: Essential Residues Required for Acetyl Transfer to Arylamines.
Biochem.J., 390, 2005
|
|
