6Q8E
 
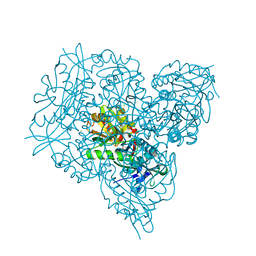 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain-amino-acid aminotransferase, CHLORIDE ION | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6ZHK
 
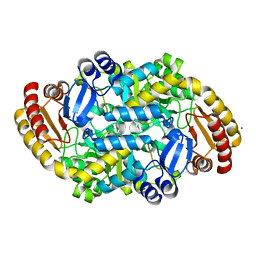 | | Crystal structure of adenosylmethionine-8-amino-7-oxononanoate aminotransferase from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, MAGNESIUM ION | | Authors: | Boyko, K.M, Nikolaeva, T.N, Stekhanova, T.N, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2020-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure of Thermostable D-Amino Acid Transaminase from the Archaeon Methanocaldococcus jannaschii DSM 2661
Crystallography Reports, 2021
|
|
3TN7
 
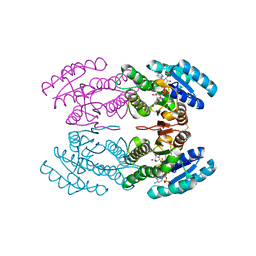 | | Crystal structure of short-chain alcohol dehydrogenase from hyperthermophilic archaeon Thermococcus sibiricus complexed with 5-hydroxy-NADP | | Descriptor: | 5-hydroxy-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, Short-chain alcohol dehydrogenase | | Authors: | Boyko, K.M, Polyakov, K.M, Bezsudnova, E.Y, Stekhanova, T.N, Gumerov, V.M, Mardanov, A.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | Deposit date: | 2011-09-01 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insight into the molecular basis of polyextremophilicity of short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Thermococcus sibiricus.
Biochimie, 94, 2012
|
|
6THQ
 
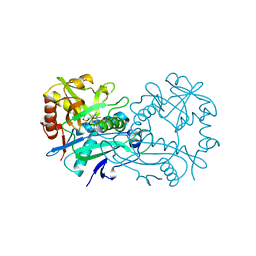 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis with norvaline | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-PENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2019-11-21 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-Dimensional Structure of Branched-Chain Amino Acid Transaminase from Thermoproteus uzoniensis in Complex with L-Norvaline
Crystallography Reports, 2020
|
|
9J32
 
 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus mutant N174K | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER | | Authors: | Ilyasov, I.O, Minyaev, M.E, Rakitina, T.V, Bakunova, A.K, Matyuta, I.O, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein of unknown function from variovorax paradoxus with amino acid substitution n174k is able to form schiff base with plp molecule
Kristallografiya, 69, 2024
|
|
3RR5
 
 | | DNA ligase from the archaeon Thermococcus sp. 1519 | | Descriptor: | DNA ligase, MAGNESIUM ION | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Popov, V.O, Polyakov, K.M, Ravin, N.V, Shabalin, I.G, Skryabin, K.G, Stekhanova, T.N, Kovalchuk, M.V. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.018 Å) | | Cite: | ATP-dependent DNA ligase from Thermococcus sp. 1519 displays a new arrangement of the OB-fold domain.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7P7X
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (holo form). | | Descriptor: | ACETATE ION, Aminotransferase class IV, PHOSPHATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis : Biochemical and Structural Insights into the New Enzyme.
Molecules, 26, 2021
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
7NEA
 
 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum (M3 mutant). | | Descriptor: | Branched-chain-amino-acid aminotransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Petrova, T, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Suplatov, D.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2021-02-03 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the residues in the active site of the transaminase from Thermobaculum terrenum.
Plos One, 16, 2021
|
|
7NEB
 
 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum (M4 mutant) | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Boyko, K.M, Petrova, T, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Suplatov, D.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2021-02-03 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the role of the residues in the active site of the transaminase from Thermobaculum terrenum.
Plos One, 16, 2021
|
|
4NMJ
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ at 2 A resolution | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ at 2 A resolution
To be Published
|
|
4NMK
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+)
To be Published
|
|
8RAF
 
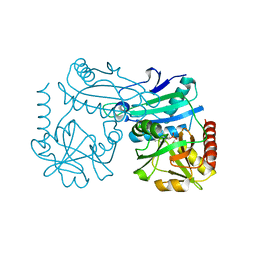 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I (holo form) | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
8RAI
 
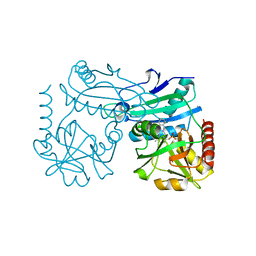 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I complexed with phenylhydrazine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
9J0U
 
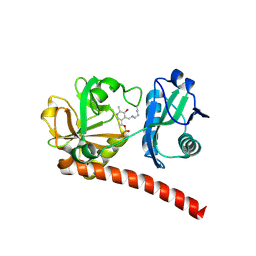 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in F 41 3 2 space group | | Descriptor: | Dat: predicted D-alanine aminotransferase | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | From Structure to Function: Analysis of the First Monomeric Pyridoxal-5'-Phosphate-Dependent Transaminase from the Bacterium Desulfobacula toluolica .
Biomolecules, 14, 2024
|
|
9J0V
 
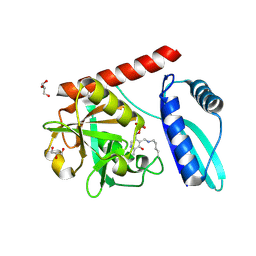 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in P 21 21 21 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dat: predicted D-alanine aminotransferase, GLYCEROL | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | From Structure to Function: Analysis of the First Monomeric Pyridoxal-5'-Phosphate-Dependent Transaminase from the Bacterium Desulfobacula toluolica .
Biomolecules, 14, 2024
|
|
8AYJ
 
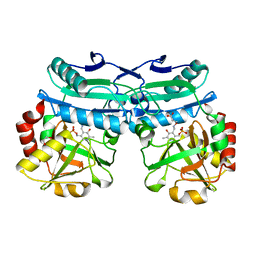 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8AYK
 
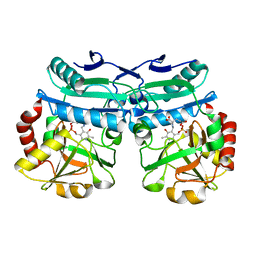 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
7Z79
 
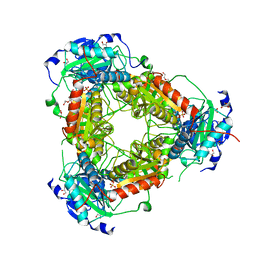 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
8AHU
 
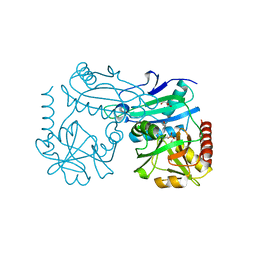 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
8AIE
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | Descriptor: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
8AHR
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
5EK6
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 complexed with NADP and isobutyraldehyde | | Descriptor: | 2-methylpropanal, Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Polyakov, K.M, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EXF
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5F2C
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-12-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
