1J7G
 
 | |
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | 分子名称: | GLUTAREDOXIN | | 著者 | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | 登録日 | 1991-10-08 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
2OUT
 
 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | 分子名称: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | 著者 | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | 登録日 | 2007-02-12 | | 公開日 | 2007-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
1PCH
 
 | |
1LBU
 
 | | HYDROLASE METALLO (ZN) DD-PEPTIDASE | | 分子名称: | MURAMOYL-PENTAPEPTIDE CARBOXYPEPTIDASE, ZINC ION | | 著者 | Charlier, P, Wery, J.-P, Dideberg, O, Frere, J.-M. | | 登録日 | 1996-03-16 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Streptomyces Albus G D-Ala-A-Ala Carboxypeptidase
Handbook of Metalloproteins, 3, 2004
|
|
2BMI
 
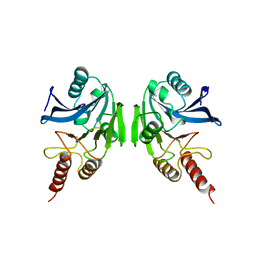 | | METALLO-BETA-LACTAMASE | | 分子名称: | PROTEIN (CLASS B BETA-LACTAMASE), SODIUM ION, ZINC ION | | 著者 | Carfi, A, Duee, E, Dideberg, O. | | 登録日 | 1998-09-17 | | 公開日 | 1998-09-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of the ZnII beta-lactamase from Bacteroides fragilis in an orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3CA8
 
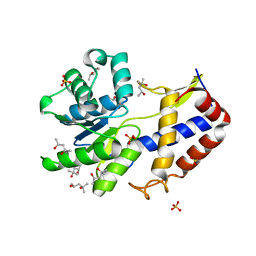 | | Crystal structure of Escherichia coli YdcF, an S-adenosyl-L-methionine utilizing enzyme | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein ydcF, SULFATE ION | | 著者 | Lim, K, Chao, K, Lehmann, C, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2008-02-19 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes.
Proteins, 72, 2008
|
|
1QME
 
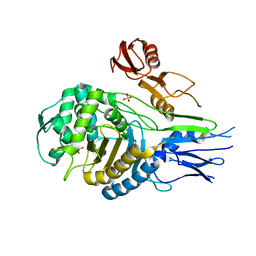 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | 分子名称: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | 著者 | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | 登録日 | 1999-09-28 | | 公開日 | 2000-05-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1DXK
 
 | |
3US1
 
 | |
3B8I
 
 | |
2XD1
 
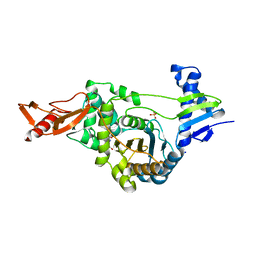 | | ACTIVE SITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS | | 分子名称: | CEFOTAXIME, C3' cleaved, open, ... | | 著者 | Macheboeuf, P, Di Guilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | 登録日 | 2010-04-28 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1EEH
 
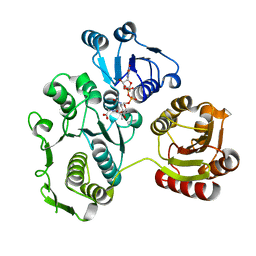 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | 分子名称: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | 著者 | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | 分子名称: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | 著者 | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | 登録日 | 1999-09-28 | | 公開日 | 2000-05-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
3ZOF
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound benzene-1,4-diol | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, benzene-1,4-diol | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
1PIO
 
 | |
3ZOE
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound p-hydroxybenzaldehyde | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, P-HYDROXYBENZALDEHYDE | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3FA4
 
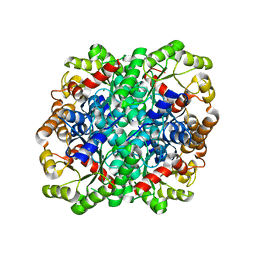 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, triclinic crystal form | | 分子名称: | 2,3-dimethylmalate lyase, MAGNESIUM ION | | 著者 | Narayanan, B.C, Herzberg, O. | | 登録日 | 2008-11-14 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
1M1B
 
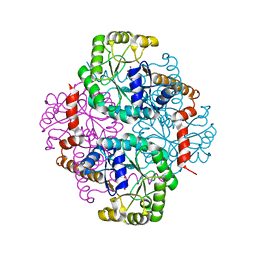 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | 分子名称: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | 著者 | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | 登録日 | 2002-06-18 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
3ZOD
 
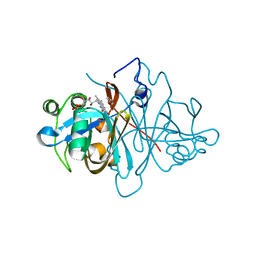 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound benzene-1,4-diol | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, benzene-1,4-diol | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
1SPH
 
 | |
3ZOC
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound p-hydroxybenzaldehyde | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, P-HYDROXYBENZALDEHYDE | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
2DIK
 
 | | R337A MUTANT OF PYRUVATE PHOSPHATE DIKINASE | | 分子名称: | PROTEIN (PYRUVATE PHOSPHATE DIKINASE), SULFATE ION | | 著者 | Huang, K, Herzberg, O. | | 登録日 | 1998-09-03 | | 公開日 | 1999-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase.
Biochemistry, 37, 1998
|
|
4OJ6
 
 | |
1M85
 
 | | Structure of Proteus mirabilis catalase for the native form | | 分子名称: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | 著者 | Gouet, P, Jouve, H.-M, Dideberg, O. | | 登録日 | 2002-07-24 | | 公開日 | 2002-08-14 | | 最終更新日 | 2014-04-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of Proteus mirabilis PR catalase with and without bound NADPH.
J.Mol.Biol., 249, 1995
|
|
