7ZRD
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | 分子名称: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
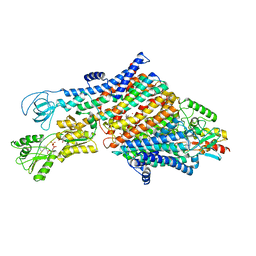 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
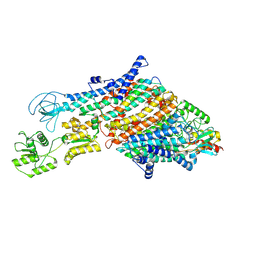 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRI
 
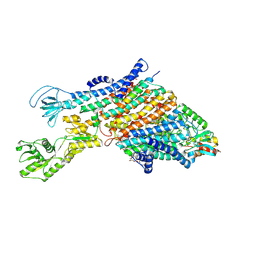 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
1AOL
 
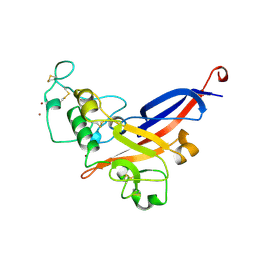 | | FRIEND MURINE LEUKEMIA VIRUS RECEPTOR-BINDING DOMAIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GP70, ZINC ION | | 著者 | Fass, D, Davey, R.A, Hamson, C.A, Kim, P.S, Cunningham, J.M, Berger, J.M. | | 登録日 | 1997-07-08 | | 公開日 | 1997-10-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution.
Science, 277, 1997
|
|
4XGC
 
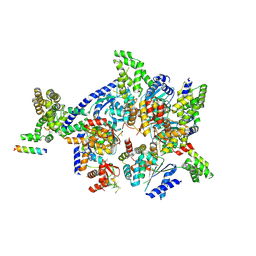 | | Crystal structure of the eukaryotic origin recognition complex | | 分子名称: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | 著者 | Bleichert, F, Botchan, M.R, Berger, J.M. | | 登録日 | 2014-12-30 | | 公開日 | 2015-04-01 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
4WMG
 
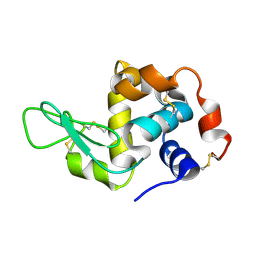 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | 分子名称: | Lysozyme C | | 著者 | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | 登録日 | 2014-10-08 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8TXR
 
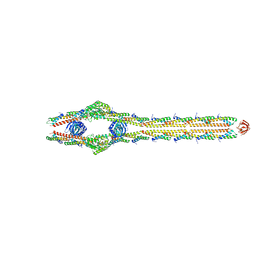 | | E. coli ExoVII(H238A) | | 分子名称: | Exodeoxyribonuclease 7 large subunit, Exodeoxyribonuclease 7 small subunit | | 著者 | Liu, C, Berger, J.M. | | 登録日 | 2023-08-24 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of Escherichia coli exonuclease VII.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5HE9
 
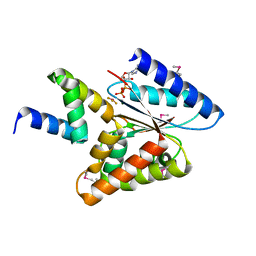 | |
5HE8
 
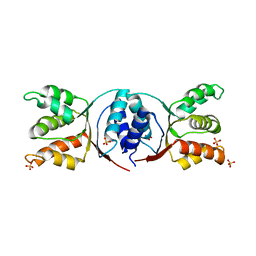 | |
1ERI
 
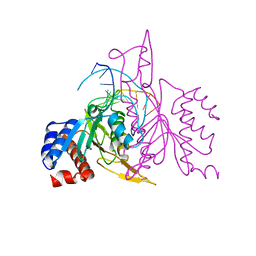 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | 分子名称: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | 著者 | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | 登録日 | 1994-05-18 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
1BJT
 
 | |
1L8Q
 
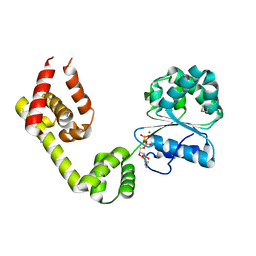 | |
1XIP
 
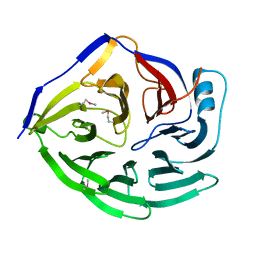 | |
2A8V
 
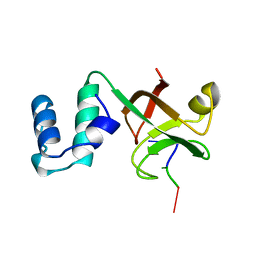 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | 分子名称: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | 著者 | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | 登録日 | 1998-11-08 | | 公開日 | 1999-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
3FK3
 
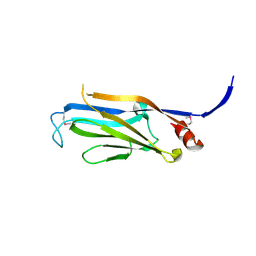 | | Structure of the Yeats Domain, Yaf9 | | 分子名称: | Protein AF-9 homolog | | 著者 | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | 登録日 | 2008-12-15 | | 公開日 | 2009-10-27 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4TMA
 
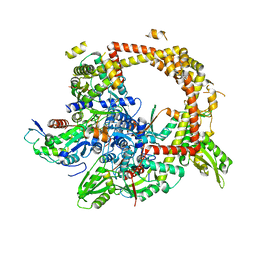 | | Crystal structure of gyrase bound to its inhibitor YacG | | 分子名称: | DNA gyrase inhibitor YacG, DNA gyrase subunit A, DNA gyrase subunit B, ... | | 著者 | Vos, S.M, Lyubimov, A.Y, Hershey, D.M, Schoeffler, A.J, Berger, J.M. | | 登録日 | 2014-05-31 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Direct control of type IIA topoisomerase activity by a chromosomally encoded regulatory protein.
Genes Dev., 28, 2014
|
|
4Z98
 
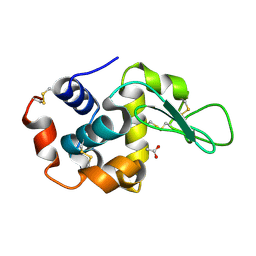 | | Crystal Structure of Hen Egg White Lysozyme using Serial X-ray Diffraction Data Collection | | 分子名称: | ACETATE ION, Lysozyme C | | 著者 | Murray, T.D, Lyubimov, A.Y, Ogata, C.M, Uervirojnangkoorn, M, Brunger, A.T, Berger, J.M. | | 登録日 | 2015-04-10 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A high-transparency, micro-patternable chip for X-ray diffraction analysis of microcrystals under native growth conditions.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5BTG
 
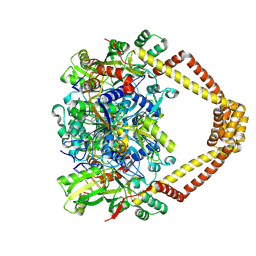 | | Crystal structure of a topoisomerase II complex | | 分子名称: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | 著者 | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | 登録日 | 2015-06-03 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CFP
 
 | | Crystal structure of anemone STING (Nematostella vectensis) 'humanized' F276K in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p]' | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.066 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5CFO
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'rotated' open conformation | | 分子名称: | Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5CFL
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p] | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CITRATE ANION, Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
