1B7V
 
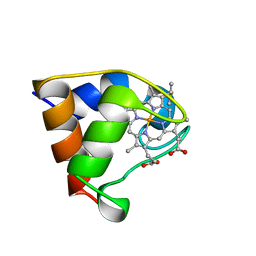 | | Structure of the C-553 cytochrome from Bacillus pasteruii to 1.7 A resolution | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C-553) | | Authors: | Gonzalez, A, Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S. | | Deposit date: | 1999-01-22 | | Release date: | 2000-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
6FRW
 
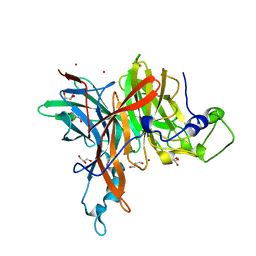 | | X-ray structure of the levansucrase from Erwinia tasmaniensis | | Descriptor: | GLYCEROL, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | Authors: | Polsinelli, I, Salomone-Stagni, M, Caliandro, R, Demitri, N, Benini, S. | | Deposit date: | 2018-02-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Comparison of the Levansucrase from the epiphyte Erwinia tasmaniensis vs its homologue from the phytopathogen Erwinia amylovora.
Int. J. Biol. Macromol., 127, 2019
|
|
6HQZ
 
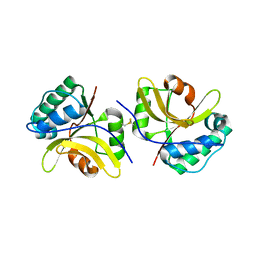 | | Crystal structure of the type III effector protein AvrRpt2 from Erwinia amylovora, a C70 family cysteine protease | | Descriptor: | AvrRpt2 | | Authors: | Bartho, J.D, Demitri, N, Wuerges, J, Benini, S. | | Deposit date: | 2018-09-25 | | Release date: | 2019-04-10 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Erwinia amylovora AvrRpt2 provides insight into protein maturation and induced resistance to fire blight by Malus × robusta 5.
J.Struct.Biol., 206, 2019
|
|
8C86
 
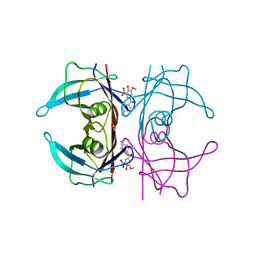 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 2 | | Descriptor: | (2,4-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
8C85
 
 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 1 | | Descriptor: | (3,5-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
7OEQ
 
 | |
6YUX
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) ternary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6YSB
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) apo form | | Descriptor: | 2-alkenal reductase (NADP(+)-dependent)-like, SULFATE ION | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6YTZ
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Double Bond Reductase, ... | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
5FR7
 
 | | Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
5FRK
 
 | | SeMet crystal structure of Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-18 | | Release date: | 2017-02-15 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
4D48
 
 | | Crystal Structure of glucose-1-phosphate uridylyltransferase GalU from Erwinia amylovora. | | Descriptor: | GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE | | Authors: | Toccafondi, M, Wuerges, J, Cianci, M, Benini, S. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Glucose-1-phosphate uridylyltransferase from Erwinia amylovora: Activity, structure and substrate specificity.
Biochim. Biophys. Acta, 1865, 2017
|
|
4D47
 
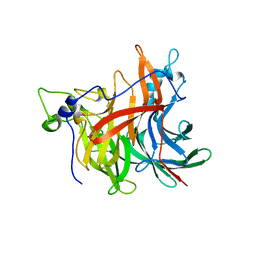 | | X-ray structure of the levansucrase from Erwinia amylovora | | Descriptor: | LEVANSUCRASE, alpha-D-glucopyranose, beta-D-fructofuranose | | Authors: | Wuerges, J, Caputi, L, Cianci, M, Benini, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | The Crystal Structure of Erwinia Amylovora Levansucrase Provides a Snapshot of the Products of Sucrose Hydrolysis Trapped Into the Active Site.
J.Struct.Biol., 191, 2015
|
|
5O3Z
 
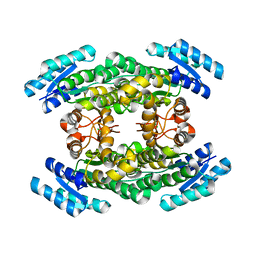 | | Crystal structure of Sorbitol-6-Phosphate 2-dehydrogenase SrlD from Erwinia amylovora | | Descriptor: | CHLORIDE ION, Sorbitol-6-phosphate dehydrogenase | | Authors: | Salomone-Stagni, M, Bartho, J.D, Bellini, D, Walsh, M.A, Benini, S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and functional analysis of Erwinia amylovora SrlD. The first crystal structure of a sorbitol-6-phosphate 2-dehydrogenase.
J.Struct.Biol., 203, 2018
|
|
3ZQM
 
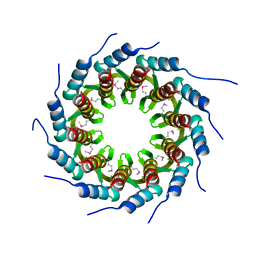 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5O7O
 
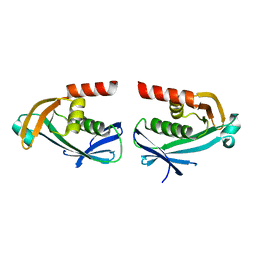 | | The crystal structure of DfoC, the desferrioxamine biosynthetic pathway acetyltransferase/Non-Ribosomal Peptide Synthetase (NRPS)-Independent Siderophore (NIS) from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | Desferrioxamine siderophore biosynthesis protein dfoC | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O5C
 
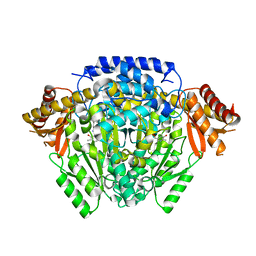 | | The crystal structure of DfoJ, the desferrioxamine biosynthetic pathway lysine decarboxylase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O8R
 
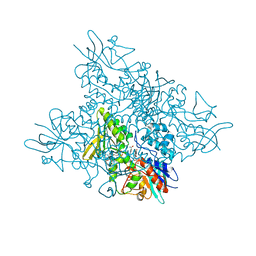 | | The crystal structure of DfoA bound to FAD and NADP; the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O8P
 
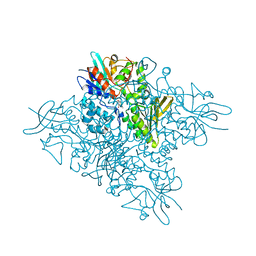 | | The crystal structure of DfoA bound to FAD, the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J.Struct.Biol., 202, 2018
|
|
3ZQN
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 2) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQO
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 3) | | Descriptor: | POTASSIUM ION, TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQQ
 
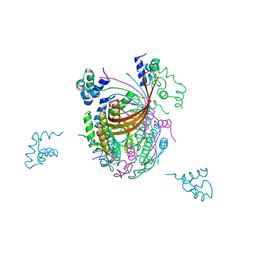 | | Crystal structure of the full-length small terminase from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQP
 
 | | Crystal structure of the small terminase oligomerization domain from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
