1YHK
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase | | Descriptor: | SULFATE ION, farnesyl pyrophosphate synthase | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
1YHL
 
 | | Structure of the complex of Trypanosoma cruzi farnesyl diphosphate synthase with risedronate, dmapp and mg+2 | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
3U6I
 
 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 58a | | Descriptor: | Hepatocyte growth factor receptor, N-{3-fluoro-4-[(7-methoxyquinolin-4-yl)oxy]phenyl}-1-[(2R)-2-hydroxypropyl]-5-methyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
2GT4
 
 | | Crystal Structure of the Y103F mutant of the GDP-mannose mannosyl hydrolase in complex with GDP-mannose and MG+2 | | Descriptor: | GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, Mildvan, A.S, Amzel, L.A. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
2GT2
 
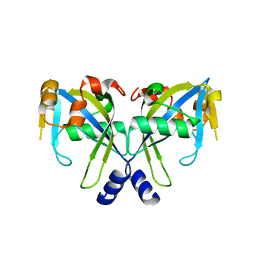 | | Structure of the E. coli GDP-mannose mannosyl hydrolase | | Descriptor: | GDP-mannose mannosyl hydrolase | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, MIldvan, A.S, Amzel, L.M. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
3U6H
 
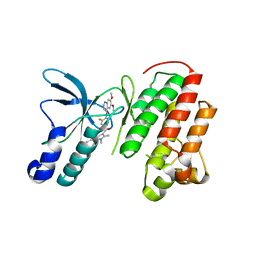 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | Descriptor: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3NLS
 
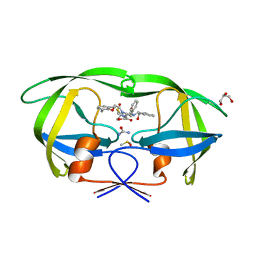 | | Crystal Structure of HIV-1 Protease in Complex with KNI-10772 | | Descriptor: | (4R)-3-[(2R,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Gabelli, S.B, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2010-06-21 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HIV-1 Protease in Complex with KNI-10772
To be Published
|
|
2O1C
 
 | |
1GA7
 
 | | CRYSTAL STRUCTURE OF THE ADP-RIBOSE PYROPHOSPHATASE IN COMPLEX WITH GD+3 | | Descriptor: | GADOLINIUM ION, HYPOTHETICAL 23.7 KDA PROTEIN IN ICC-TOLC INTERGENIC REGION | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2000-11-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.
Nat.Struct.Biol., 8, 2001
|
|
1RMD
 
 | | RAG1 DIMERIZATION DOMAIN | | Descriptor: | RAG1, ZINC ION | | Authors: | Bellon, S.F, Rodgers, K.K, Schatz, D.G, Coleman, J.E, Steitz, T.A. | | Deposit date: | 1997-01-10 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
5T3Q
 
 | |
1G0S
 
 | |
1G9Q
 
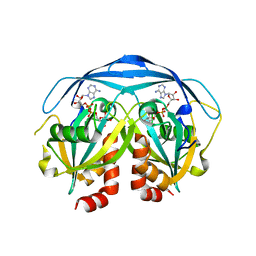 | | COMPLEX STRUCTURE OF THE ADPR-ASE AND ITS SUBSTRATE ADP-RIBOSE | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, HYPOTHETICAL 23.7 KDA PROTEIN IN ICC-TOLC INTERGENIC REGION | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.
Nat.Struct.Biol., 8, 2001
|
|
1KHZ
 
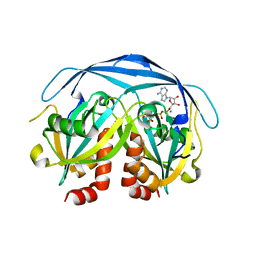 | | Structure of the ADPR-ase in complex with AMPCPR and Mg | | Descriptor: | ADP-ribose pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2001-12-01 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase.
Biochemistry, 41, 2002
|
|
2XVC
 
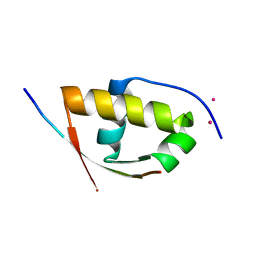 | | Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division | | Descriptor: | CADMIUM ION, CDVA, SSO0911, ... | | Authors: | Samson, R.Y, Obita, T, Hodgson, B, Shaw, M.K, Chong, P.L, Williams, R.L, Bell, S.D. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular and Structural Basis of Escrt-III Recruitment to Membranes During Archaeal Cell Division.
Mol.Cell, 41, 2011
|
|
8DYB
 
 | |
8E5J
 
 | | The crystal structure of 4-n-butylbenzoic acid bound CYP199A4 | | Descriptor: | 4-butylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Doherty, D.Z, Bell, S.G, Bruning, J.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the Factors which Result in Cytochrome P450 Catalyzed Desaturation Versus Hydroxylation.
Chem Asian J, 17, 2022
|
|
4OV1
 
 | | The crystal structure of a novel electron transfer ferredoxin from R. palustris HaA2 | | Descriptor: | FE3-S4 CLUSTER, Putative ferredoxin | | Authors: | Zhouw, W.H, Zhang, T, Zhang, A.L, Bell, S.G, Wong, L.-L. | | Deposit date: | 2014-02-19 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | The structure of a novel electron-transfer ferredoxin from Rhodopseudomonas palustris HaA2 which contains a histidine residue in its iron-sulfur cluster-binding motif.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KDY
 
 | |
3HF2
 
 | | Crystal structure of the I401P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2009-05-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Highly Active Single-Mutation Variant of P450(BM3) (CYP102A1)
Chembiochem, 10, 2009
|
|
4V8S
 
 | | Archaeal RNAP-DNA binary complex at 4.32Ang | | Descriptor: | 5'-D(*AP*TP*AP*GP*AP*GP*TP*AP*TP*AP*AP*GP*AP*TP *AP*G)-3', 5'-D(*TP*CP*TP*TP*AP*TP*AP*CP*TP*CP*TP*AP*TP*CP)-3', DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-07-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.323 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
6DCD
 
 | |
2FRZ
 
 | |
8GLZ
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid. Crystal was initially co-crystallised with 4-methoxybenzoic acid and soaked with 4 mM hydrogen peroxide | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
8GM2
 
 | |
