4S29
 
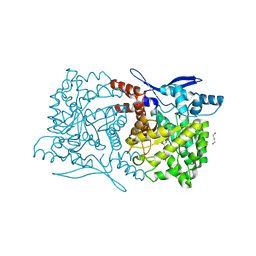 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide and Fe | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, FE (II) ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
3FPZ
 
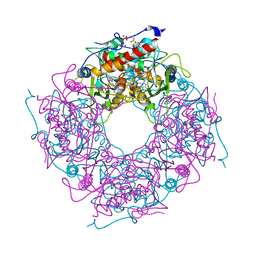 | | Saccharomyces cerevisiae THI4p is a suicide thiamin thiazole synthase | | Descriptor: | ADENOSINE DIPHOSPHATE 5-(BETA-ETHYL)-4-METHYL-THIAZOLE-2-CARBOXYLIC ACID, SULFATE ION, Thiazole biosynthetic enzyme | | Authors: | Bale, S, Chatterjee, A, Dorrestein, P.C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase.
Nature, 478, 2011
|
|
4ESX
 
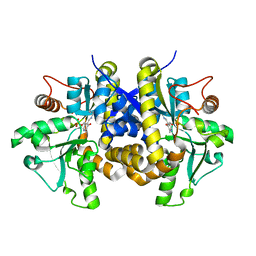 | | Crystal structure of C. albicans Thi5 complexed with PLP | | Descriptor: | Pyrimidine biosynthesis enzyme THI13 | | Authors: | Huang, S, Fenwick, M.K, Zhang, Y, Lai, R, Hazra, A, Rajashankar, K, Philmus, B, Kinsland, C, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thiamin pyrimidine biosynthesis in Candida albicans : a remarkable reaction between histidine and pyridoxal phosphate.
J.Am.Chem.Soc., 134, 2012
|
|
4ESW
 
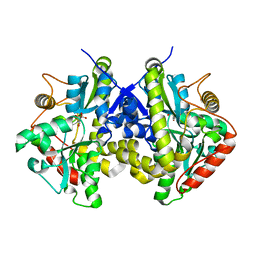 | | Crystal structure of C. albicans Thi5 H66G mutant | | Descriptor: | CITRIC ACID, Pyrimidine biosynthesis enzyme THI13 | | Authors: | Fenwick, M.K, Huang, S, Zhang, Y, Lai, R, Hazra, A, Rajashankar, K, Philmus, B, Kinsland, C, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thiamin pyrimidine biosynthesis in Candida albicans : a remarkable reaction between histidine and pyridoxal phosphate.
J.Am.Chem.Soc., 134, 2012
|
|
4KYS
 
 | | Clostridium botulinum thiaminase I in complex with thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Sikowitz, M.D, Shome, B, Zhang, Y, Begley, T.P, Ealick, S.E. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure of a Clostridium botulinum C143S Thiaminase I/Thiamin Complex Reveals Active Site Architecture.
Biochemistry, 52, 2013
|
|
4JEL
 
 | | Structure of MilB Streptomyces rimofaciens CMP N-glycosidase | | Descriptor: | CMP/hydroxymethyl CMP hydrolase, SULFATE ION | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
4JEN
 
 | | Structure of Clostridium botulinum CMP N-glycosidase, BcmB | | Descriptor: | CMP N-GLYCOSIDASE, PHOSPHATE ION | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
4JEM
 
 | | Crystal structure of MilB complexed with cytidine 5'-monophosphate | | Descriptor: | CMP/hydroxymethyl CMP hydrolase, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
1YFX
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans complexed with 4-chloro-3-hydroxyanthranilic acid and NO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate-3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
1YFW
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans complexed with 4-chloro-3-hydroxyanthranilic acid and O2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate-3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
1YAK
 
 | | Complex of Bacillus subtilis TenA with 4-amino-2-methyl-5-hydroxymethylpyrimidine | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, Transcriptional activator tenA | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
1YAD
 
 | | Structure of TenI from Bacillus subtilis | | Descriptor: | Regulatory protein tenI, SULFATE ION, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
1YAF
 
 | | Structure of TenA from Bacillus subtilis | | Descriptor: | Transcriptional activator tenA | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
1YFY
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans complexed with 3-hydroxyanthranilic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate-3,4-dioxygenase, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
1YFU
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate-3,4-dioxygenase, CHLORIDE ION, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
3GMC
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase with substrate bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-6-methylpyridine-3-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
4GIL
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase/Linear Pseudouridine 5'-Phosphate Adduct | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, pseudouridine 5'-phosphate, ... | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.539 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
3GMB
 
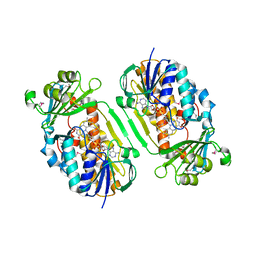 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
4GIJ
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Sulfate | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, SULFATE ION | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIM
 
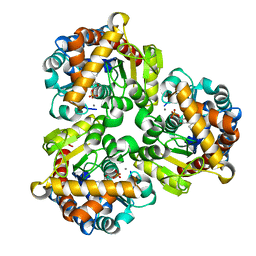 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Pseudouridine 5'-phosphate | | Descriptor: | MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, Pseudouridine-5'-phosphate glycosidase | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIK
 
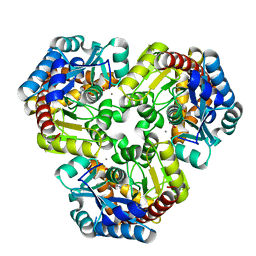 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase/Linear R5P Adduct | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, RIBOSE-5-PHOSPHATE | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4HHE
 
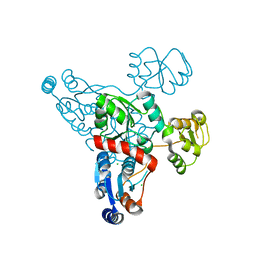 | | Quinolinate synthase from Pyrococcus furiosus | | Descriptor: | CHLORIDE ION, Quinolinate synthase A | | Authors: | Soriano, E.V, Zhang, Y, Settembre, E.C, Colabroy, K, Sanders, J.M, Dorrestein, P.C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-10-09 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Active-site models for complexes of quinolinate synthase with substrates and intermediates.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3IX1
 
 | | Periplasmic N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding protein from Bacillus halodurans | | Descriptor: | N-[(4-amino-2-methylpyrimidin-5-yl)methyl]formamide, N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding protein | | Authors: | Bale, S, Rajashankar, K.R, Perry, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-09-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | HMP Binding Protein ThiY and HMP-P Synthase THI5 Are Structural Homologues.
Biochemistry, 49, 2010
|
|
3EPM
 
 | | Crystal structure of Caulobacter crescentus ThiC | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SULFATE ION, Thiamine biosynthesis protein thiC, ... | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3DWG
 
 | | Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis | | Descriptor: | 9.5 kDa culture filtrate antigen cfp10A, Cysteine synthase B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jurgenson, C.T, Burns, K.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis.
Biochemistry, 47, 2008
|
|
