1CM5
 
 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | 分子名称: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-14 | | 公開日 | 1999-12-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
3PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | 分子名称: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-14 | | 公開日 | 2000-05-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | 分子名称: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | 分子名称: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
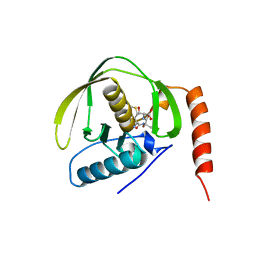 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSZ
 
 | | PEPTIDE DEFORMYLASE AS FE2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | 分子名称: | FE (III) ION, NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | 分子名称: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
4TLW
 
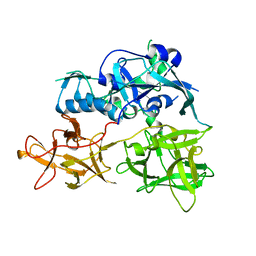 | | CARDS TOXIN, FULL-LENGTH | | 分子名称: | ADP-ribosylating toxin CARDS | | 著者 | Becker, A, GALALELDEEN, A, Taylor, A.B, Hart, P.J. | | 登録日 | 2014-05-30 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | 分子名称: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-26 | | 公開日 | 1999-12-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1H18
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate | | 分子名称: | FORMATE ACETYLTRANSFERASE 1, L-TREITOL, PYRUVIC ACID, ... | | 著者 | Becker, A, Kabsch, W. | | 登録日 | 2002-07-04 | | 公開日 | 2002-11-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1ICJ
 
 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | 分子名称: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-03-12 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1H16
 
 | |
1H17
 
 | |
1OKT
 
 | | X-ray Structure of Glutathione S-Transferase from the Malarial Parasite Plasmodium falciparum | | 分子名称: | FORMIC ACID, GLUTATHIONE S-TRANSFERASE | | 著者 | Fritz-Wolf, K, Becker, A, Rahlfs, s, Harwaldt, P, Schirmer, R.H, Kabsch, W, Becker, K. | | 登録日 | 2003-07-29 | | 公開日 | 2003-11-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-Ray Structure of Glutathione S-Transferase from the Malarial Parasite Plasmodium Falciparum
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3QQL
 
 | | CDK2 in complex with inhibitor L3 | | 分子名称: | (5R)-5-(2-methylbutan-2-yl)-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-02-15 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R8P
 
 | | CDK2 in complex with inhibitor NSK-MC1-6 | | 分子名称: | (5R)-5-propyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R8L
 
 | | CDK2 in complex with inhibitor L3-4 | | 分子名称: | (5R)-5-tert-butyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QZF
 
 | | CDK2 in complex with inhibitor JWS-6-52 | | 分子名称: | 2-(4,6-diamino-1,3,5-triazin-2-yl)benzene-1,4-diol, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-06 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QXO
 
 | | CDK2 in complex with inhibitor KVR-1-84 | | 分子名称: | 1,2-ETHANEDIOL, 5-nitro-2-[(4-sulfamoylbenzyl)amino]benzamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-02 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QWJ
 
 | | CDK2 in complex with inhibitor KVR-1-142 | | 分子名称: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-chloro-5-nitrobenzamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-02-28 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QX2
 
 | | CDK2 in complex with inhibitor KVR-1-190 | | 分子名称: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-[(2-hydroxyethyl)amino]-5-nitrobenzamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-01 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QZI
 
 | | CDK2 in complex with inhibitor KVR-1-126 | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-5-nitro-2-[(pyrimidin-5-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-06 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R28
 
 | | CDK2 in complex with inhibitor KVR-1-140 | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-5-nitro-2-[(3,4,5-trifluorobenzyl)amino]benzamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-14 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R73
 
 | | CDK2 in complex with inhibitor KVR-1-164 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(3-aminopropyl)amino]-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-22 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
