1KFL
 
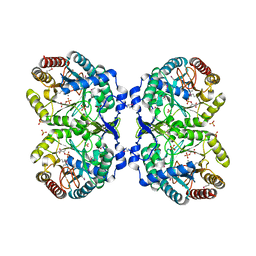 | | Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from E.coli complexed with Mn2+, PEP, and Phe | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Shumilin, I.A, Zhao, C, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2001-11-21 | | Release date: | 2002-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase alters the coordination of both substrates.
J.Mol.Biol., 320, 2002
|
|
4C2H
 
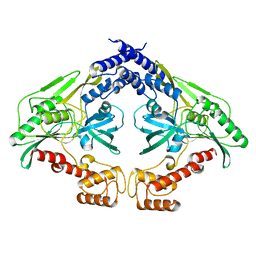 | | Crystal structure of the CtpB(V118Y) mutant | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2F
 
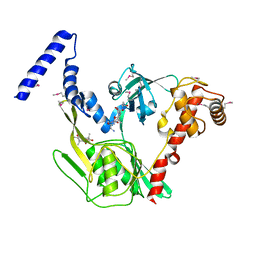 | | Crystal structure of the CtpB R168A mutant present in an active conformation | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
1N8F
 
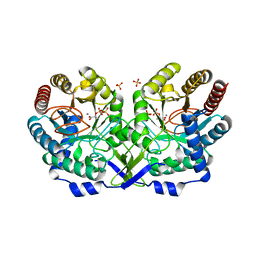 | | Crystal structure of E24Q mutant of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Escherichia Coli in complex with Mn2+ and PEP | | Descriptor: | DAHP Synthetase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2002-11-20 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The High-Resolution Structure of 3-Deoxy-D-arabino-heptulosonate-7-phosphate
Synthase Reveals a Twist in the Plane of Bound Phosphoenolpyruvate
Biochemistry, 42, 2003
|
|
4HPK
 
 | | Crystal structure of Clostridium histolyticum colg collagenase collagen-binding domain 3B at 1.35 Angstrom resolution in presence of calcium nitrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase, ... | | Authors: | Philominathan, S.T.L, Wilson, J.J, Bauer, R, Matsushita, O, Sakon, J. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
4BNB
 
 | |
4BN7
 
 | |
1XQ8
 
 | |
1QR7
 
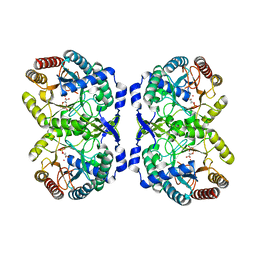 | | CRYSTAL STRUCTURE OF PHENYLALANINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE FROM ESCHERICHIA COLI COMPLEXED WITH PB2+ AND PEP | | Descriptor: | LEAD (II) ION, PHENYLALANINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Kretsinger, R.H, Bauerle, R.H. | | Deposit date: | 1999-06-18 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1NPS
 
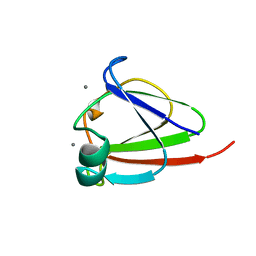 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
4BN8
 
 | |
2JLE
 
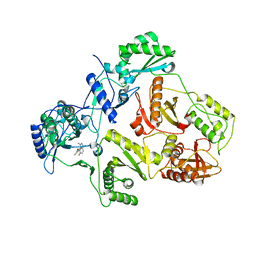 | | Novel indazole nnrtis created using molecular template hybridization based on crystallographic overlays | | Descriptor: | 5-[(5-fluoro-3-methyl-1H-indazol-4-yl)oxy]benzene-1,3-dicarbonitrile, REVERSE TRANSCRIPTASE/RNASEH | | Authors: | Jones, L.H, Allan, G, Barba, O, Burt, C, Corbau, R, Dupont, T, Irving, S, Mowbray, C.E, Phillips, C, Swain, N.A, Webster, R, Westby, M. | | Deposit date: | 2008-09-08 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Indazole Non-Nucleoside Reverse Transcriptase Inhibitors Using Molecular Hybridization Based on Crystallographic Overlays.
J.Med.Chem., 52, 2009
|
|
4JRW
 
 | | Crystal structure of Clostridium histolyticum colg collagenase PKD domain 2 at 1.6 Angstrom resolution | | Descriptor: | BROMIDE ION, Collagenase | | Authors: | Sakon, J, Philominathan, S.T.L, Gann, S, Bauer, R, Matsushita, O. | | Deposit date: | 2013-03-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3B4P
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, complex with 2-(cyclohexylamino)benzoic acid | | Descriptor: | 2-(cyclohexylamino)benzoic acid, ACETATE ION, AZIDE ION, ... | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3B4O
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
1QDL
 
 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | Authors: | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1999-05-20 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6FOE
 
 | | BaxB01 Fab fragment | | Descriptor: | Fab BaxB01 heavy chain, Fab BaxB01 light chain | | Authors: | Hollerweger, J, Schinagl, A, Kerschbaumer, R.J, Scheiflinger, F, Thiele, M, Goettig, P, Brandstetter, H. | | Deposit date: | 2018-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Biochemistry, 57, 2018
|
|
