7T43
 
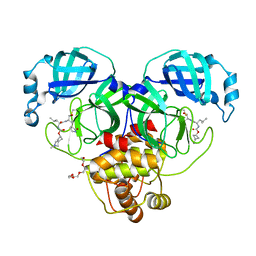 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4A
 
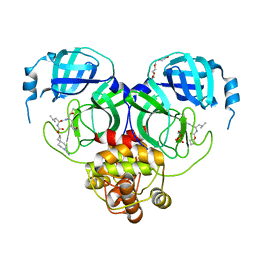 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 11c | | Descriptor: | (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Z
 
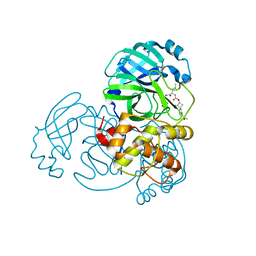 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T45
 
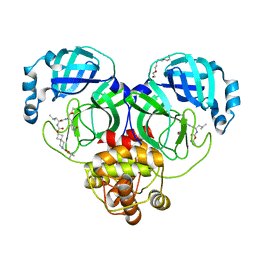 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 7c | | Descriptor: | (1S,2S)-2-{[N-({[7-(tert-butoxycarbonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Kankanamalage, A.C.G, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T48
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T44
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 4c | | Descriptor: | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7SNW
 
 | | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576 | | Descriptor: | 2-(methoxycarbonyl)thiophene-3-sulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576
To be published
|
|
7SNY
 
 | | 2.10A Resolution Structure of NanoBiT Complementation Reporter Large Subunit LgBiT | | Descriptor: | Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.10A Resolution Structure of NanoBiT Complementation Reporter Large Subunit LgBiT
To be published
|
|
7SNT
 
 | | 2.20A Resolution Structure of NanoLuc Luciferase with Bound Substrate Analog 3-methoxy-furimazine | | Descriptor: | (4S)-8-benzyl-2-[(furan-2-yl)methyl]-3-methoxy-6-phenylimidazo[1,2-a]pyrazine, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Unch, J, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.20A Resolution Structure of NanoLuc Luciferase with Bound Substrate Analog 3-methoxy-furimazine
To be published
|
|
7SNR
 
 | | 2.00A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, CITRATE ANION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00A Resolution Structure of NanoLuc Luciferase
To be published
|
|
7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
7SNX
 
 | | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits | | Descriptor: | GLYCEROL, Oplophorus-luciferin 2-monooxygenase catalytic subunit, Oplophorus-luciferin 2-monooxygenase catalytic subunit: C-terminal Peptide (11-mer) | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits
To be published
|
|
7TQ7
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ8
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ4
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ2
 
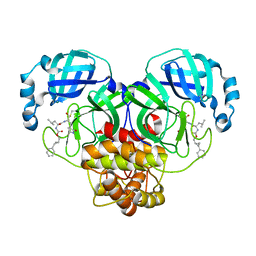 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ3
 
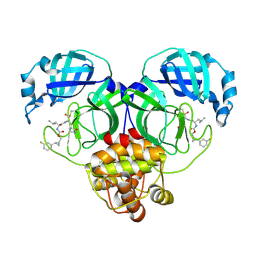 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
9BK3
 
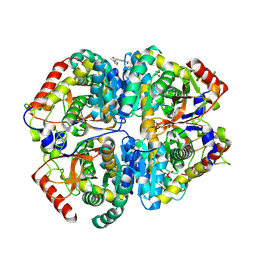 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BTS
 
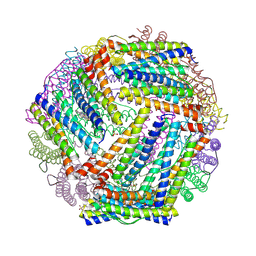 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
9BK2
 
 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
5UE0
 
 | | 1.90 A resolution structure of CT622 C-terminal domain from Chlamydia trachomatis | | Descriptor: | CT622 protein, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2016-12-29 | | Release date: | 2018-01-10 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Loss of Expression of a Single Type 3 Effector (CT622) Strongly ReducesChlamydia trachomatisInfectivity and Growth.
Front Cell Infect Microbiol, 8, 2018
|
|
4EAN
 
 | | 1.75A resolution structure of indole bound beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-galactosidase, CHLORIDE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
4E5D
 
 | | 2.2A resolution structure of a firefly luciferase-benzothiazole inhibitor complex | | Descriptor: | 2-(2-fluorophenyl)-6-methoxy-1,3-benzothiazole, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Throne, N, Shen, M, Auld, D.S, Inglese, J. | | Deposit date: | 2012-03-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.
Chem.Biol., 19, 2012
|
|
