1LSO
 
 | |
1IL0
 
 | | X-RAY CRYSTAL STRUCTURE OF THE E170Q MUTANT OF HUMAN L-3-HYDROXYACYL-COA DEHYDROGENASE | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, ACETOACETYL-COENZYME A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2001-05-07 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glutamate 170 of human l-3-hydroxyacyl-CoA dehydrogenase is required for proper orientation of the catalytic histidine and structural integrity of the enzyme.
J.Biol.Chem., 276, 2001
|
|
2QMC
 
 | |
2QM6
 
 | |
2HDH
 
 | |
1M76
 
 | |
1M75
 
 | |
1LSJ
 
 | |
1F14
 
 | |
1F0Y
 
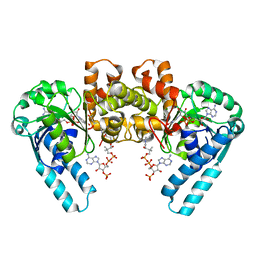 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH ACETOACETYL-COA AND NAD+ | | Descriptor: | ACETOACETYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F17
 
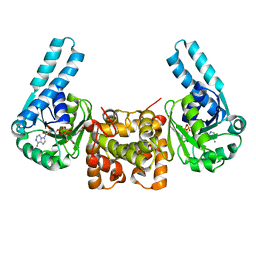 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F12
 
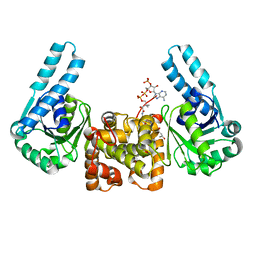 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH 3-HYDROXYBUTYRYL-COA | | Descriptor: | 3-HYDROXYBUTANOYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
3LVV
 
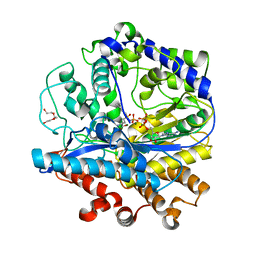 | | BSO-inhibited ScGCL | | Descriptor: | (2S)-2-amino-4-(S-butyl-N-phosphonosulfonimidoyl)butanoic acid, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Barycki, J.J, Biterova, E.I. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for feedback and pharmacological inhibition of Saccharomyces cerevisiae glutamate cysteine ligase.
J.Biol.Chem., 285, 2010
|
|
3HDH
 
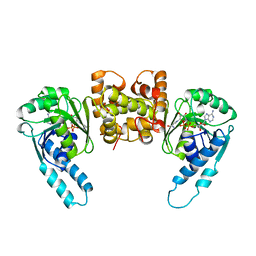 | | PIG HEART SHORT CHAIN L-3-HYDROXYACYL COA DEHYDROGENASE REVISITED: SEQUENCE ANALYSIS AND CRYSTAL STRUCTURE DETERMINATION | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) | | Authors: | Barycki, J.J, O'Brien, L.K, Birktoft, J.J, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 1999-04-13 | | Release date: | 1999-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pig heart short chain L-3-hydroxyacyl-CoA dehydrogenase revisited: sequence analysis and crystal structure determination.
Protein Sci., 8, 1999
|
|
3HAD
 
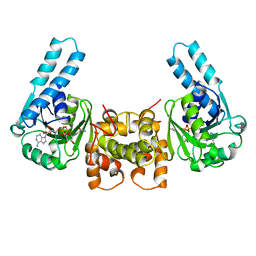 | |
1ZDL
 
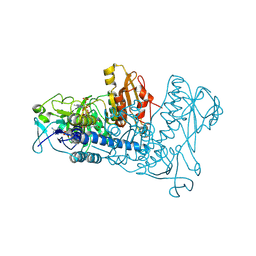 | | Crystal Structure of Mouse Thioredoxin Reductase Type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 2, ... | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-04-14 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZKQ
 
 | | Crystal structure of mouse thioredoxin reductase type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 2, mitochondrial | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-05-03 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2NQO
 
 | | Crystal Structure of Helicobacter pylori gamma-Glutamyltranspeptidase | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Boanca, G, Sand, A, Okada, T, Suzuki, H, Kumagai, H, Fukuyama, K, Barycki, J.J. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad.
J.Biol.Chem., 282, 2007
|
|
3LVW
 
 | | Glutathione-inhibited ScGCL | | Descriptor: | GLUTATHIONE, Glutamate--cysteine ligase, TRIETHYLENE GLYCOL | | Authors: | Biterova, E.I, Barycki, J.J. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for feedback and pharmacological inhibition of Saccharomyces cerevisiae glutamate cysteine ligase.
J.Biol.Chem., 285, 2010
|
|
3FNM
 
 | | Crystal structure of acivicin-inhibited gamma-glutamyltranspeptidase reveals critical roles for its C-terminus in autoprocessing and catalysis | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, Gamma-glutamyltranspeptidase (Ggt) Large subunit, Gamma-glutamyltranspeptidase (Ggt) Small subunit | | Authors: | Williams, K, Cullati, S, Sand, A, Biterova, E.I, Barycki, J.J. | | Deposit date: | 2008-12-25 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Acivicin-Inhibited gamma-Glutamyltranspeptidase Reveals Critical Roles for Its C-Terminus in Autoprocessing and Catalysis.
Biochemistry, 48, 2009
|
|
3IG8
 
 | |
3IG5
 
 | |
