7VOI
 
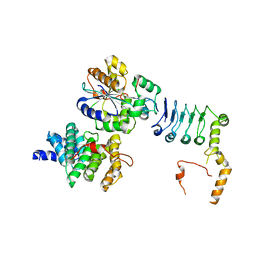 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | Authors: | Bartlam, M, Zhang, Q. | | Deposit date: | 2021-10-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.38 Å) | | Cite: | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
1VE7
 
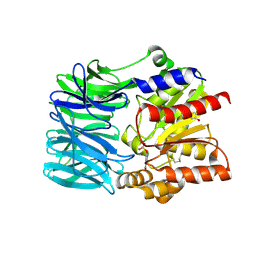 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
1VE6
 
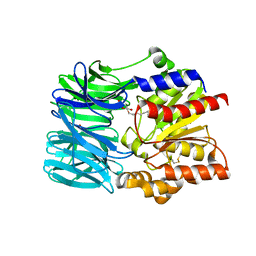 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
8Y1J
 
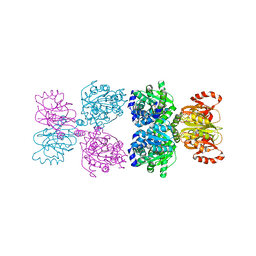 | |
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
8K8L
 
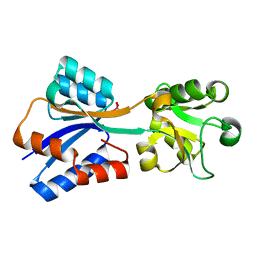 | |
8K8K
 
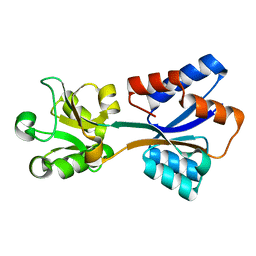 | | Structure of Klebsiella pneumonia ModA | | Descriptor: | Molybdate transporter periplasmic protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 681, 2023
|
|
7BVD
 
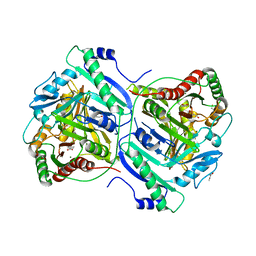 | | Anthranilate synthase component I (TrpE)[Mycolicibacterium smegmatis] | | Descriptor: | Anthranilate synthase component 1, BENZOIC ACID, GLYCEROL, ... | | Authors: | Chen, Y, Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit I of the anthranilate synthase complex of Mycolicibacterium smegmatis
Biochem.Biophys.Res.Commun., 527, 2020
|
|
1C76
 
 | | STAPHYLOKINASE (SAK) MONOMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Staphylokinase Dimer Offers New Clue to Reduction of Immunogenicity
To be published
|
|
1C77
 
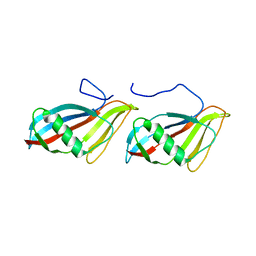 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C78
 
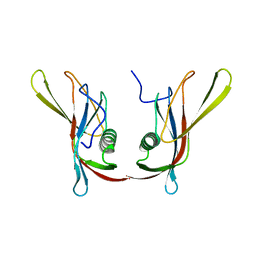 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C79
 
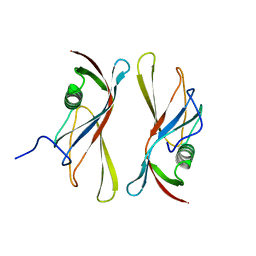 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
5DV2
 
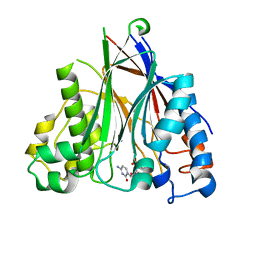 | |
5DV4
 
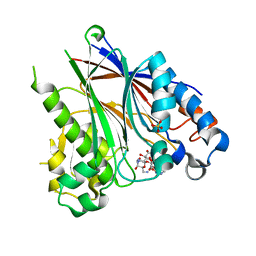 | |
4MFI
 
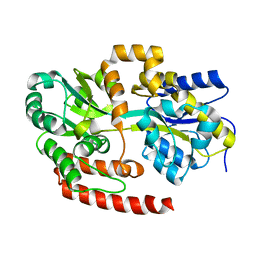 | | Crystal structure of Mycobacterium tuberculosis UgpB | | Descriptor: | Sn-glycerol-3-phosphate ABC transporter substrate-binding protein UspB | | Authors: | Jiang, D, Bartlam, M, Rao, Z. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine
Febs J., 281, 2014
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6D
 
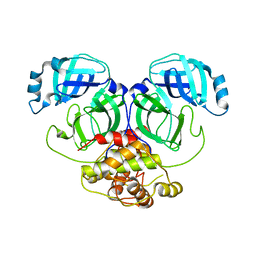 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
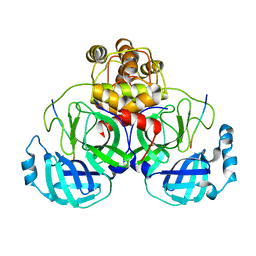 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2QC3
 
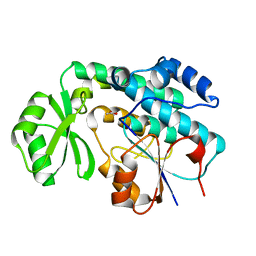 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
1P9E
 
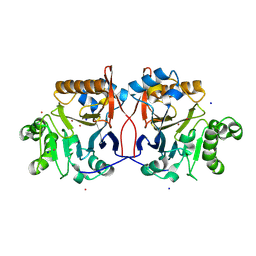 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
1GHE
 
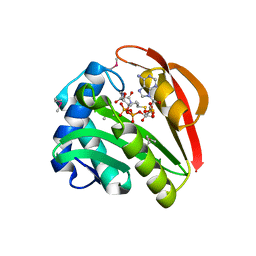 | | CRYSTAL STRUCTURE OF TABTOXIN RESISTANCE PROTEIN COMPLEXED WITH AN ACYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, ACETYLTRANSFERASE | | Authors: | He, H, Ding, Y, Bartlam, M, Sun, F, Le, Y, Qin, X, Tang, H, Zhang, R, Joachimiak, A, Liu, Y, Zhao, N, Rao, Z. | | Deposit date: | 2000-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Tabtoxin Resistance Protein Complexed with Acetyl
Coenzyme A Reveals the Mechanism for beta-Lactam Acetylation
J.Mol.Biol., 325, 2003
|
|
5WQN
 
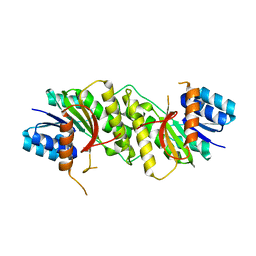 | |
5WQO
 
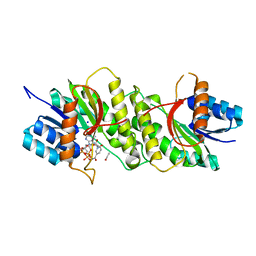 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WQM
 
 | |
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
