5A1A
 
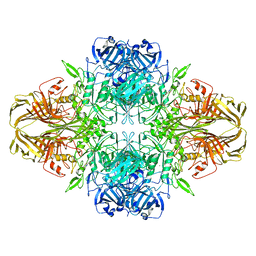 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | 著者 | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | 登録日 | 2015-04-29 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
4CC8
 
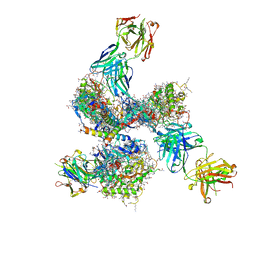 | | Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy | | 分子名称: | GP120, GP41, MONOCLONAL ANTIBODY VRC03 FAB HEAVY CHAIN, ... | | 著者 | Bartesaghi, A, Merk, A, Borgnia, M.J, Milne, J.L.S, Subramaniam, S. | | 登録日 | 2013-10-18 | | 公開日 | 2013-10-30 | | 最終更新日 | 2018-01-10 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Prefusion Structure of Trimeric HIV-1 Envelope Glycoprotein Determined by Cryo-Electron Microscopy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2YNJ
 
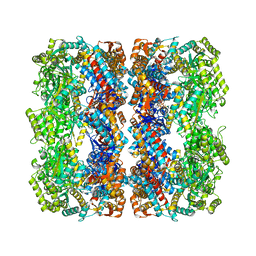 | | GroEL at sub-nanometer resolution by Constrained Single Particle Tomography | | 分子名称: | 60 KDA CHAPERONIN, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Bartesaghi, A, Lecumberry, F, Sapiro, G, Subramaniam, S. | | 登録日 | 2012-10-15 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography
Structure, 20, 2012
|
|
3J7H
 
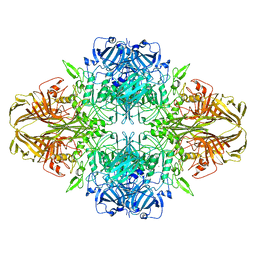 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | 分子名称: | Beta-galactosidase, MAGNESIUM ION | | 著者 | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5K0Z
 
 | | Cryo-EM structure of lactate dehydrogenase (LDH) in inhibitor-bound state | | 分子名称: | L-lactate dehydrogenase B chain | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
4UQJ
 
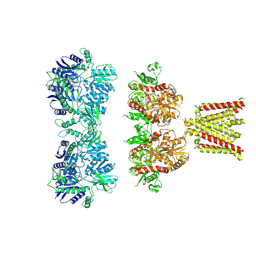 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | 分子名称: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-24 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (10.4 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQK
 
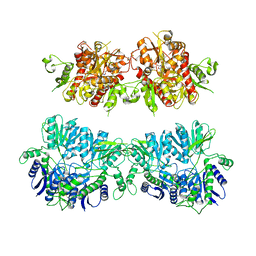 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-24 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (16.4 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQ6
 
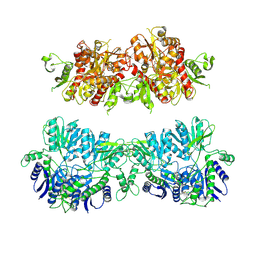 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | 分子名称: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQQ
 
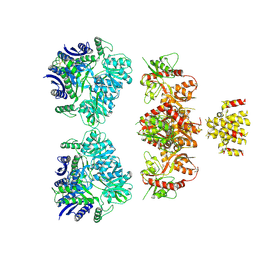 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | 分子名称: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-24 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | 分子名称: | Regulatory protein NPR1, ZINC ION | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAD
 
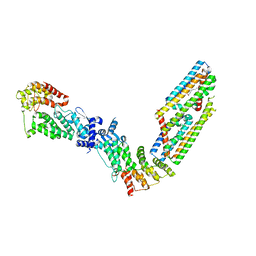 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | 分子名称: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | 著者 | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | 登録日 | 2021-12-20 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | 分子名称: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | 著者 | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | 登録日 | 2021-12-20 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
5K10
 
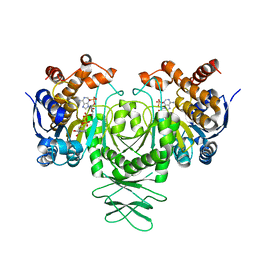 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K11
 
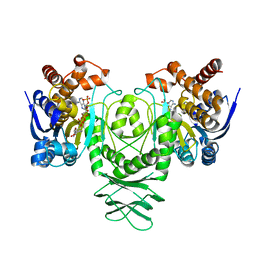 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in inhibitor-bound state | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K12
 
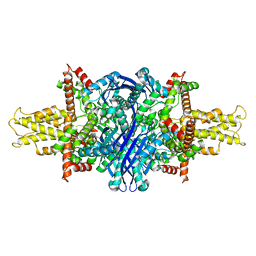 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | 分子名称: | Glutamate dehydrogenase 1, mitochondrial | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (1.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
6XIR
 
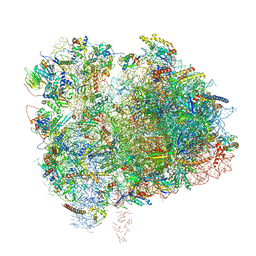 | | Cryo-EM Structure of K63 Ubiquitinated Yeast Translocating Ribosome under Oxidative Stress | | 分子名称: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhou, Y, Bartesaghi, A, Silva, G.M. | | 登録日 | 2020-06-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XIQ
 
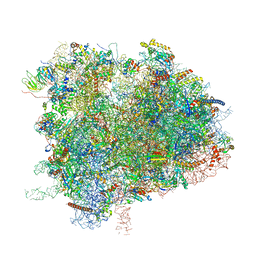 | | Cryo-EM Structure of K63R Ubiquitin Mutant Ribosome under Oxidative Stress | | 分子名称: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhou, Y, Bartesaghi, A, Silva, G.M. | | 登録日 | 2020-06-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3DNO
 
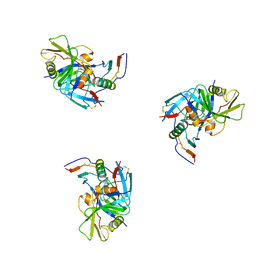 | | Molecular structure for the HIV-1 gp120 trimer in the CD4-bound state | | 分子名称: | HIV-1 envelope glycoprotein gp120 | | 著者 | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | 登録日 | 2008-07-02 | | 公開日 | 2008-08-19 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DNN
 
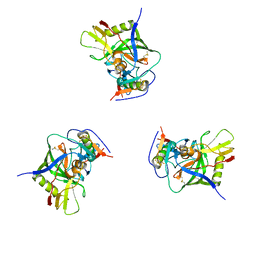 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | 分子名称: | HIV-1 envelope glycoprotein gp120 | | 著者 | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | 登録日 | 2008-07-02 | | 公開日 | 2008-08-19 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | 分子名称: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EBK
 
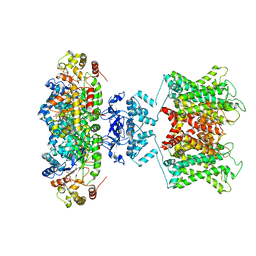 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6CVM
 
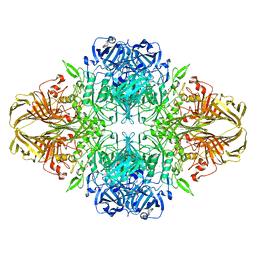 | | Atomic resolution cryo-EM structure of beta-galactosidase | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | 著者 | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | 登録日 | 2018-03-28 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (1.9 Å) | | 主引用文献 | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
3DNL
 
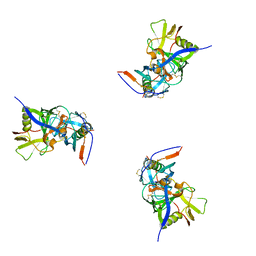 | | Molecular structure for the HIV-1 gp120 trimer in the b12-bound state | | 分子名称: | HIV-1 envelope glycoprotein gp120 | | 著者 | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | 登録日 | 2008-07-02 | | 公開日 | 2008-08-19 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
6EBL
 
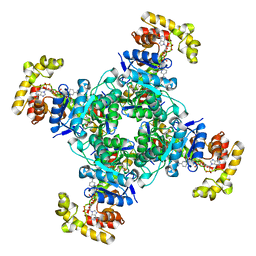 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
3JD2
 
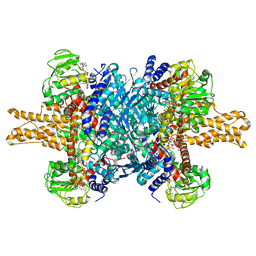 | | Glutamate dehydrogenase in complex with NADH, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
