1Z1M
 
 | | NMR structure of unliganded MDM2 | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Uhrinova, S, Uhrin, D, Powers, H, Watt, K, Zheleva, D, Fischer, P, McInnes, C, Barlow, P.N. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Free MDM2 N-terminal Domain Reveals Conformational Adjustments that Accompany p53-binding
J.Mol.Biol., 350, 2005
|
|
1VVE
 
 | |
1VVD
 
 | |
1PPQ
 
 | | NMR structure of 16th module of Immune Adherence Receptor, Cr1 (Cd35) | | Descriptor: | Complement receptor type 1 | | Authors: | O'Leary, J.M, Bromek, K, Black, G.M, Uhrinova, S, Schmitz, C, Krych, M, Atkinson, J.P, Uhrin, D, Barlow, P.N. | | Deposit date: | 2003-06-17 | | Release date: | 2004-05-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces.
Protein Sci., 13, 2004
|
|
1XWE
 
 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
1QK9
 
 | | The solution structure of the domain from MeCP2 that binds to methylated DNA | | Descriptor: | METHYL-CPG-BINDING PROTEIN 2 | | Authors: | Wakefield, R.I.D, Smith, B.O, Nan, X, Free, A, Soteriou, A, Uhrin, D, Bird, A.P, Barlow, P.N. | | Deposit date: | 1999-07-12 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Domain from Mecp2 that Binds to Methylated DNA
J.Mol.Biol., 291, 1999
|
|
2A55
 
 | | Solution structure of the two N-terminal CCP modules of C4b-binding protein (C4BP) alpha-chain. | | Descriptor: | C4b-binding protein | | Authors: | Jenkins, H.T, Mark, L, Ball, G, Lindahl, G, Uhrin, D, Blom, A.M, Barlow, P.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Human C4b-binding Protein, Structural Basis for Interaction with Streptococcal M Protein, a Major Bacterial Virulence Factor
J.Biol.Chem., 281, 2006
|
|
1NWV
 
 | | SOLUTION STRUCTURE OF A FUNCTIONALLY ACTIVE COMPONENT OF DECAY ACCELERATING FACTOR | | Descriptor: | Complement decay-accelerating factor | | Authors: | Uhrinova, S, Lin, F, Ball, G, Bromek, K, Uhrin, D, Medof, M.E, Barlow, P.N. | | Deposit date: | 2003-02-07 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a functionally active fragment of decay-accelerating factor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RXL
 
 | | Solution structure of the engineered protein Afae-dsc | | Descriptor: | Afimbrial adhesin AFA-III | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, du Merle, L, Barlow, P.N, Medof, M.E, Smith, R.A, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-18 | | Release date: | 2005-01-11 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | An atomic resolution model for assembly, architecture, and function of the Dr adhesins.
Mol.Cell, 15, 2004
|
|
1USZ
 
 | | SeMet AfaE-3 adhesin from Escherichia Coli | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, CHLORIDE ION, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1USQ
 
 | | Complex of E. Coli DraE adhesin with Chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, ... | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-11-27 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1UT1
 
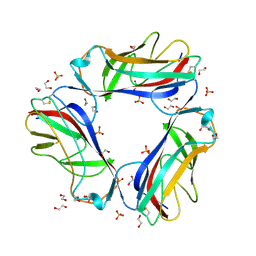 | | DraE adhesin from Escherichia Coli | | Descriptor: | 1,2-ETHANEDIOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1UT2
 
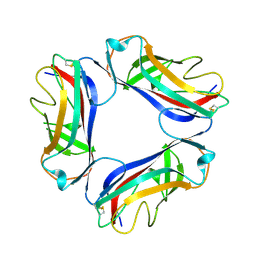 | | AfaE-3 adhesin from Escherichia Coli | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1SS2
 
 | | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 cis conformer | | Descriptor: | Gamma-aminobutyric acid type B receptor, subunit 1 | | Authors: | Blein, S, Uhrin, D, Smith, B.O, White, J.H, Barlow, P.N. | | Deposit date: | 2004-03-23 | | Release date: | 2004-10-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the complement control protein (CCP) modules of GABA(B) receptor 1a: only one of the two CCP modules is compactly folded.
J.Biol.Chem., 279, 2004
|
|
1SRZ
 
 | | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 trans conformer | | Descriptor: | Gamma-aminobutyric acid type B receptor, subunit 1 | | Authors: | Blein, S, Uhrin, D, Smith, B.O, White, J.H, Barlow, P.N. | | Deposit date: | 2004-03-23 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Complement Control Protein (CCP) Modules of GABAB Receptor 1a: ONLY ONE OF THE TWO CCP MODULES IS COMPACTLY FOLDED
J.Biol.Chem., 279, 2004
|
|
1E5G
 
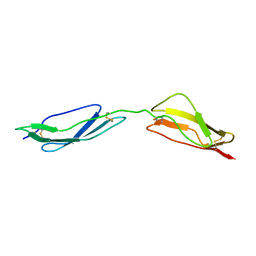 | | Solution structure of central CP module pair of a pox virus complement inhibitor | | Descriptor: | COMPLEMENT CONTROL PROTEIN C3 | | Authors: | Henderson, C.E, Bromek, K, Mullin, N.P, Smith, B.O, Uhrin, D, Barlow, P.N. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-31 | | Last modified: | 2013-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Central Ccp Module Pair of a Poxvirus Complement Control Protein
J.Mol.Biol., 307, 2001
|
|
1GKN
 
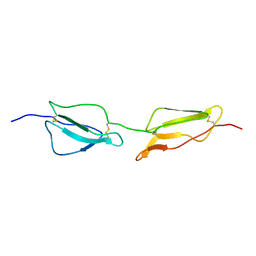 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
1H67
 
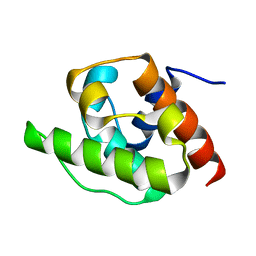 | | NMR Structure of the CH Domain of Calponin | | Descriptor: | CALPONIN ALPHA | | Authors: | Bramham, J, Smith, B.O, Uhrin, D, Barlow, P.N, Winder, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2002-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Calponin Ch Domain and Fitting to the 3D-Helical Reconstruction of F-Actin:Calponin.
Structure, 10, 2002
|
|
