6X84
 
 | |
3DYR
 
 | |
5KCI
 
 | | Crystal Structure of HTC1 | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein YPL067C, ... | | Authors: | Martin, R.M, Horowitz, S, Koepnick, B, Cooper, S, Flatten, J, Rogawski, D.S, Koropatkin, N.M, Beinlich, F.R.M, Players, F, Students, U.M, Popovic, Z, Baker, D, Khatib, F, Bardwell, J.C.A. | | Deposit date: | 2016-06-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Determining crystal structures through crowdsourcing and coursework.
Nat Commun, 7, 2016
|
|
7TQR
 
 | |
7TLX
 
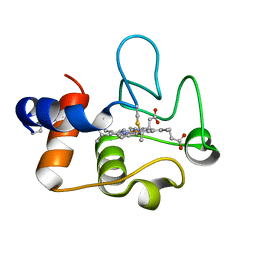 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
8DSV
 
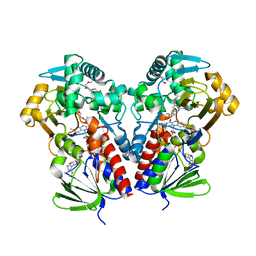 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
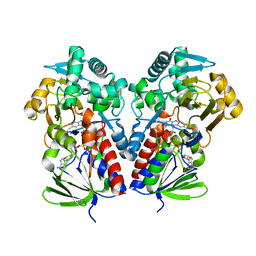 | |
8DQ8
 
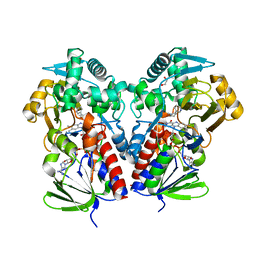 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
1DSB
 
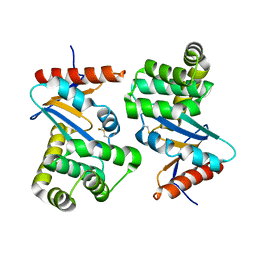 | |
1DM9
 
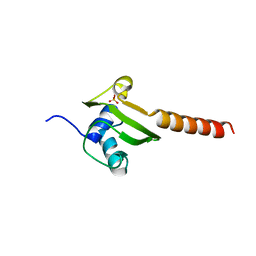 | | HEAT SHOCK PROTEIN 15 KD | | Descriptor: | HYPOTHETICAL 15.5 KD PROTEIN IN MRCA-PCKA INTERGENIC REGION, SULFATE ION | | Authors: | Staker, B.L, Korber, P, Bardwell, J.C.A, Saper, M.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hsp15 reveals a novel RNA-binding motif.
EMBO J., 19, 2000
|
|
1EIZ
 
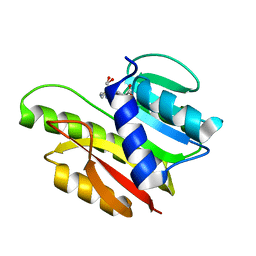 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE | | Descriptor: | FTSJ, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
1EJ0
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE, MERCURY DERIVATIVE | | Descriptor: | FTSJ, MERCURY (II) ION, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
5WO1
 
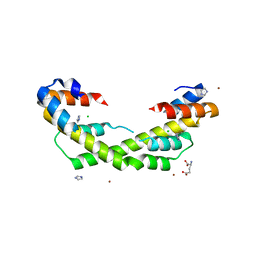 | | Chaperone Spy H96L bound to Im7 L18A L19A L37A (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, IMIDAZOLE, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO3
 
 | | Chaperone Spy bound to Im7 (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6OWX
 
 | | Spy H96L:Im7 L18pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWY
 
 | | Spy H96L:Im7 K20pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWZ
 
 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2H0G
 
 | | Crystal Structure of DsbG T200M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
2H0I
 
 | | Crystal Structure of DsbG V216M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
2H0H
 
 | | Crystal Structure of DsbG K113E mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
