3GUZ
 
 | | Structural and substrate-binding studies of pantothenate synthenate (PS)provide insights into homotropic inhibition by pantoate in PS's | | Descriptor: | PANTOATE, Pantothenate synthetase | | Authors: | Chakrabarti, K.S, Thakur, K.G, Gopal, B, Sarma, S.P. | | Deposit date: | 2009-03-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and NMR studies of pantothenate synthetase provide insights into the mechanism of homotropic inhibition by pantoate
Febs J., 277, 2010
|
|
4CMB
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | ACETATE ION, N4-cyclohexyl-5-(4-fluorophenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CM5
 
 | |
4CMK
 
 | |
4CM6
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (E)-2-amino-4-oxo-6-styryl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CMA
 
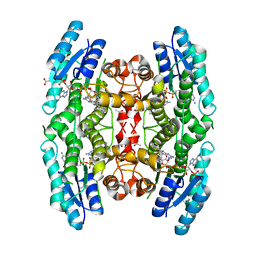 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
2QDB
 
 | |
3POK
 
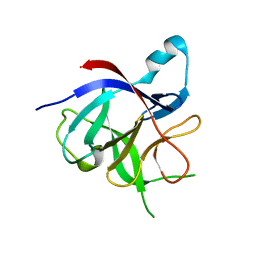 | | Interleukin-1-beta LBT L3 Mutant | | Descriptor: | Interleukin-1 beta | | Authors: | Barthelmes, K, Reynolds, A.M, Peisach, E, Jonker, H.R.A, DeNunzio, N.J, Allen, K.N, Imperiali, B, Schwalbe, H. | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering encodable lanthanide-binding tags into loop regions of proteins.
J.Am.Chem.Soc., 133, 2011
|
|
5AJ8
 
 | |
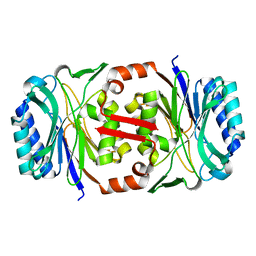
2W1V
 
 | |
1QFA
 
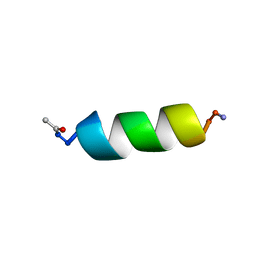 | | STRUCTURE OF A NEUROPEPTIDE Y Y2 AGONIST | | Descriptor: | PROTEIN (NEUROPEPTIDE Y) | | Authors: | Barnham, K.J, Catalfamo, F, Pallaghy, P.K, Howlett, G.J, Norton, R.S. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Helical structure and self-association in a 13 residue neuropeptide Y Y2 receptor agonist: relationship to biological activity.
Biochim.Biophys.Acta, 1435, 1999
|
|
2XOX
 
 | | Crystal structure of pteridine reductase (PTR1) from Leishmania donovani | | Descriptor: | PTERIDINE REDUCTASE, SULFATE ION | | Authors: | Barrack, K.L, Tulloch, L.B, Burke, L.A, Fyfe, P.K, Hunter, W.N. | | Deposit date: | 2010-08-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Recombinant Leishmania Donovani Pteridine Reductase Reveals a Disordered Active Site.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2RDF
 
 | |
4CQI
 
 | | Crystal structure of recombinant tubulin-binding cofactor A (TBCA) from Leishmania major | | Descriptor: | GLYCEROL, SULFATE ION, TUBULIN-BINDING COFACTOR A | | Authors: | Barrack, K.L, Fyfe, P.K, Hunter, W.N. | | Deposit date: | 2014-02-17 | | Release date: | 2014-11-26 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Tubulin-Binding Cofactor a from Leishmania Major Infers a Mode of Association During the Early Stages of Microtubule Assembly
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1OWT
 
 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
1ZEC
 
 | | NMR Solution structure of NEF1-25, 20 structures | | Descriptor: | NEF1-25 | | Authors: | Barnham, K.J, Monks, S.A, Hinds, M.G, Azad, A.A, Norton, R.S. | | Deposit date: | 1996-12-18 | | Release date: | 1998-01-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Biochemistry, 36, 1997
|
|
5EC8
 
 | |
5E9Q
 
 | |
5EIF
 
 | |
5EHG
 
 | |
5EHI
 
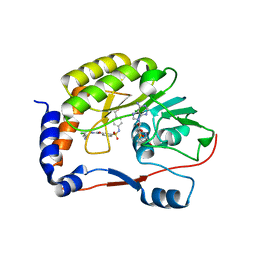 | |
5EIW
 
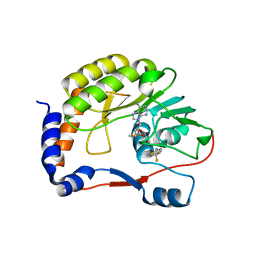 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYL METHIONINE AND FRAGMENT NB3C2 | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, NS5 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2015-10-30 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design.
Eur.J.Med.Chem., 125, 2016
|
|
5EKX
 
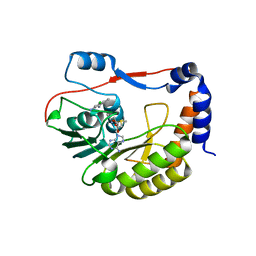 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYLMETHIONINE AND FRAGMENT NB2E11 | | Descriptor: | 4-chloro-5-methylbenzene-1,2-diamine, NS5 METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design.
Eur.J.Med.Chem., 125, 2016
|
|
4KYC
 
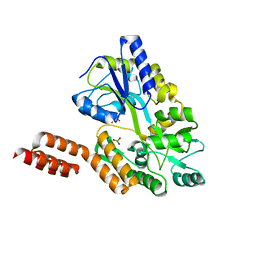 | | Structure of the C-terminal domain of the Menangle virus phosphoprotein, fused to MBP. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Maltose-binding periplasmic protein, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
4KYE
 
 | | Partial Structure of the C-terminal domain of the HPIV4B phosphoprotein, fused to MBP. | | Descriptor: | Maltose-binding periplasmic protein, Phosphoprotein, chimeric construct, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
