6OI5
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, mitochondrial | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
6OIB
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
6OIC
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfite soaked) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, Sulfide:quinone oxidoreductase, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
6OI6
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfide soaked) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, mitochondrial, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
2PEL
 
 | | PEANUT LECTIN | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEANUT LECTIN, ... | | Authors: | Banerjee, R, Das, K, Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformation, protein-carbohydrate interactions and a novel subunit association in the refined structure of peanut lectin-lactose complex.
J.Mol.Biol., 259, 1996
|
|
4DVD
 
 | |
3RX6
 
 | | Crystal structure of Polarity Suppression protein from Enterobacteria phage P4 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, MERCURY (II) ION, ... | | Authors: | Banerjee, R, Nath, S, Khamrui, S, Sen, R, Sen, U. | | Deposit date: | 2011-05-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | The first structure of polarity suppression protein, Psu from enterobacteria phage P4, reveals a novel fold and a knotted dimer
J.Biol.Chem., 287, 2012
|
|
6WH6
 
 | | Crystal structure of human sulfide quinone oxidoreductase in complex with coenzyme Q (cyanide soaked) | | Descriptor: | CYANIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, ... | | Authors: | Banerjee, R, Cho, U.S, Moon, S. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dismantling and Rebuilding the Trisulfide Cofactor Demonstrates Its Essential Role in Human Sulfide Quinone Oxidoreductase.
J.Am.Chem.Soc., 142, 2020
|
|
6X8Z
 
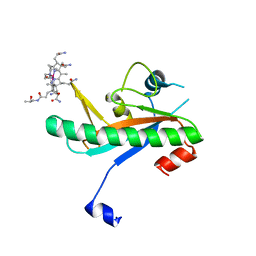 | | Crystal structure of N-truncated human B12 chaperone CblD(C262S)-thiolato-cob(III)alamin complex (108-296) | | Descriptor: | COBALAMIN, Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Mascarenhas, R, Li, Z, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interprotein Co-S Coordination Complex in the B 12 -Trafficking Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
8I0Z
 
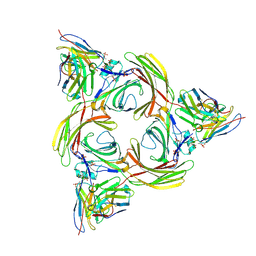 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 (Local refine) | | Descriptor: | Beta-arrestin-2, C5a anaphylatoxin chemotactic receptor 1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
7RUU
 
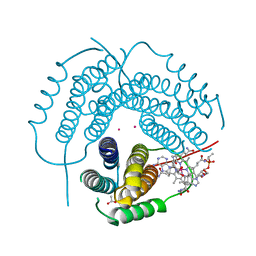 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
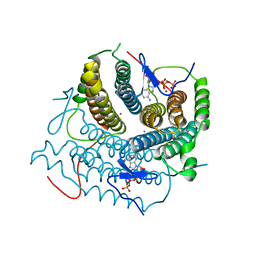 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUV
 
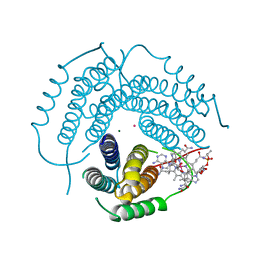 | | Structure of Human ATP:Cobalamin Adenosyltransferase E193K bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
8I0Q
 
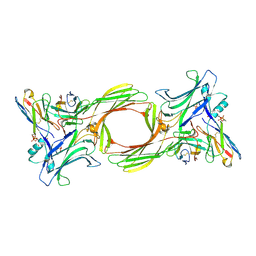 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 (Local refine) | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8I10
 
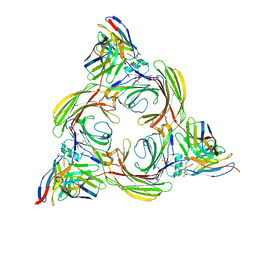 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Vasopressin V2 receptor, V2R (Local refine) | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8I9L
 
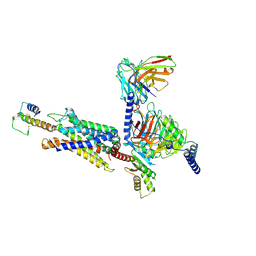 | | Structure of C3a-C3aR-Go complex (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I9S
 
 | | Structure of Apo-C3aR-Go complex (Titan) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I95
 
 | | Structure of EP54-C3aR-Go complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP54 ligand, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8IA2
 
 | | Structure of C5a bound human C5aR1 in complex with Go (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I97
 
 | | Structure of Apo-C3aR-Go complex (Glacios) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I9A
 
 | | Structure of EP54-C3aR-Gq complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP54 ligand, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
7S6Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S7H
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL2 | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Methane monooxygenase beta chain, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S6T
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit H33A | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S6R
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with H5A mutation | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
