4NTN
 
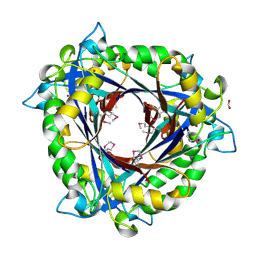 | | E.coli QueD, SeMet protein, 2A resolution | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, FORMIC ACID, ZINC ION | | Authors: | Bandarian, V, Roberts, S.A, Miles, Z.D. | | Deposit date: | 2013-12-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily.
J.Biol.Chem., 289, 2014
|
|
4NTK
 
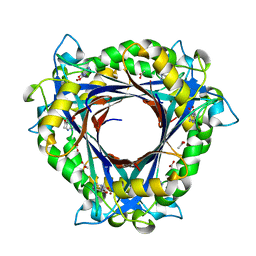 | | QueD from E. coli | | Descriptor: | 2-amino-6-[(1Z)-1,2-dihydroxyprop-1-en-1-yl]-7,8-dihydropteridin-4(3H)-one, 6-carboxy-5,6,7,8-tetrahydropterin synthase, ACETATE ION, ... | | Authors: | Bandarian, V, Roberts, S.A, Miles, Z.D. | | Deposit date: | 2013-12-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily.
J.Biol.Chem., 289, 2014
|
|
4NTM
 
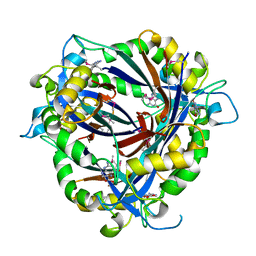 | | QueD soaked with sepiapterin (selenomethionine substituted protein) | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 6-carboxy-5,6,7,8-tetrahydropterin synthase, ZINC ION | | Authors: | Bandarian, V, Miles, Z.D, Roberts, S.A. | | Deposit date: | 2013-12-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily.
J.Biol.Chem., 289, 2014
|
|
1K98
 
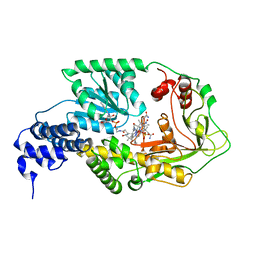 | | AdoMet complex of MetH C-terminal fragment | | Descriptor: | COBALAMIN, Methionine synthase, SULFATE ION | | Authors: | Bandarian, V, Pattridge, K.A, Lennon, B.W, Huddler, D.P, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2001-10-27 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Domain alternation switches B(12)-dependent methionine synthase to the activation conformation.
Nat.Struct.Biol., 9, 2002
|
|
1K7Y
 
 | | E. coli MetH C-terminal fragment (649-1227) | | Descriptor: | COBALAMIN, SULFATE ION, methionine synthase | | Authors: | Bandarian, V, Pattridge, K.A, Lennon, B.W, Huddler, D.P, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Domain alternation switches B(12)-dependent methionine synthase to the activation conformation.
Nat.Struct.Biol., 9, 2002
|
|
3D7J
 
 | | SCO6650, a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein SCO6650 | | Authors: | Spoonamore, J.E, Roberts, S.A, Heroux, A, Bandarian, V. | | Deposit date: | 2008-05-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2QV6
 
 | |
4NJJ
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Manganese(II) | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJG
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxypterin | | Descriptor: | 6-CARBOXYPTERIN, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJK
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 7-carboxy-7-deazaguanine, and Mg2+ | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
5T8Y
 
 | | Structure of epoxyqueuosine reductase from Bacillus subtilis with the Asp134 catalytic loop swung out of the active site. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Maiocco, S.J, Elliott, S.J, Bandarian, V, Drennan, C.L. | | Deposit date: | 2016-09-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
4NJH
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxy-5,6,7,8-tetrahydropterin | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJI
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Mg2+ | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
5D0A
 
 | | Crystal structure of epoxyqueuosine reductase with cleaved RNA stem loop | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, GLYCEROL, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Bandarian, V, Drennan, C.L. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5D08
 
 | | Crystal structure of selenomethionine-labeled epoxyqueuosine reductase | | Descriptor: | CHLORIDE ION, COBALAMIN, Epoxyqueuosine reductase, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Bandarian, V, Drennan, C.L. | | Deposit date: | 2015-08-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5D0B
 
 | | Crystal structure of epoxyqueuosine reductase with a tRNA-TYR epoxyqueuosine-modified tRNA stem loop | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, GLYCEROL, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Bandarian, V, Drennan, C.L. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
