2AF2
 
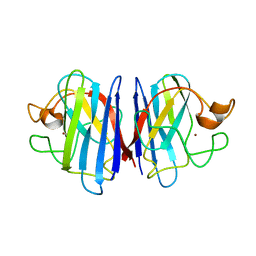 | | Solution structure of disulfide reduced and copper depleted Human Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Amelio, N, Gaggelli, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-07-25 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human SOD1 before harboring the catalytic metal: solution structure of copper-depleted, disulfide-reduced form
J.Biol.Chem., 281, 2006
|
|
2AJ1
 
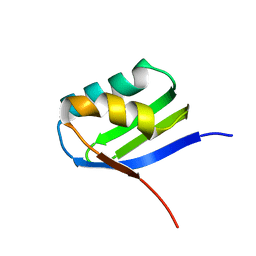 | | Solution structure of apoCadA | | Descriptor: | Probable cadmium-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.-C, Miras, R, Bal, N, Mintz, E, Catty, P, Shokes, J.E, Scott, R.A. | | Deposit date: | 2005-08-01 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for metal binding specificity: the N-terminal cadmium binding domain of the P1-type ATPase CadA
J.Mol.Biol., 356, 2006
|
|
4H2D
 
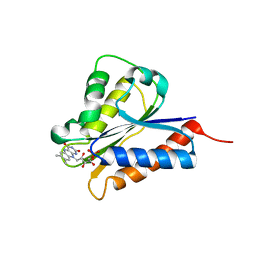 | | Crystal structure of NDOR1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-dependent diflavin oxidoreductase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mikolajczyk, M, Jaiswal, D, Winkelmann, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CJK
 
 | | Crystal structure of the adduct HAH1-Cd(II)-MNK1. | | Descriptor: | CADMIUM ION, Copper transport protein ATOX1, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Felli, I, Della-Malva, N, Pavelkova, A, Rosato, A. | | Deposit date: | 2008-03-13 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces.
Biochem.J., 422, 2009
|
|
3O55
 
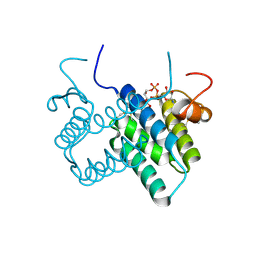 | | Crystal structure of human FAD-linked augmenter of liver regeneration (ALR) | | Descriptor: | Augmenter of liver regeneration, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A, Kallergi, E, Lionaki, E, Pozidis, C, Tokatlidis, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2M
 
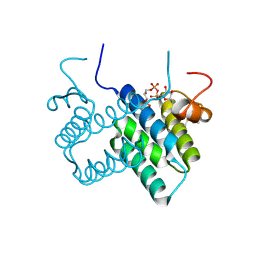 | | Crystal structure of human ALR mutant C142/145S | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-10-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An electron-transfer path through an extended disulfide relay system: the case of the redox protein ALR.
J.Am.Chem.Soc., 134, 2012
|
|
3U2L
 
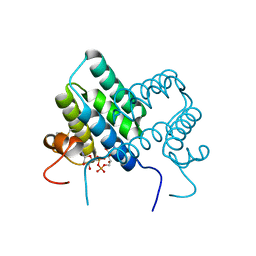 | | Crystal structure of human ALR mutant C142S. | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-10-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An electron-transfer path through an extended disulfide relay system: the case of the redox protein ALR.
J.Am.Chem.Soc., 134, 2012
|
|
2AJ0
 
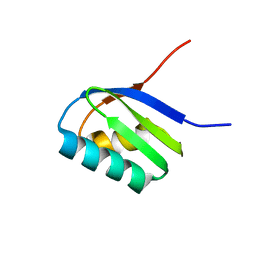 | | Solution structure of apoCadA | | Descriptor: | Probable cadmium-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.-C, Miras, R, Bal, N, Mintz, E, Catty, P, Shokes, J.E, Scott, R.A. | | Deposit date: | 2005-08-01 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for metal binding specificity: the N-terminal cadmium binding domain of the P1-type ATPase CadA
J.Mol.Biol., 356, 2006
|
|
2ROP
 
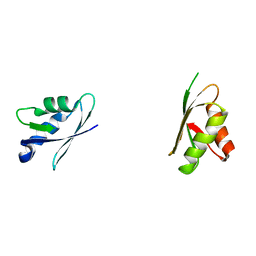 | | Solution structure of domains 3 and 4 of human ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Banci, L, Bertini, I, Cantini, F, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1
Biochemistry, 47, 2008
|
|
2RSQ
 
 | | Copper(I) loaded form of the first domain of the human copper chaperone for SOD1, CCS | | Descriptor: | COPPER (I) ION, Copper chaperone for superoxide dismutase | | Authors: | Banci, L, Bertini, I, Cantini, F, Kozyreva, T, Rubino, J.T. | | Deposit date: | 2012-05-15 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Copper(I) loaded form of the first domain of the human copper chaperone for SOD1, CCS
To be Published
|
|
2RLI
 
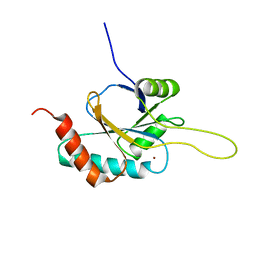 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
2RNB
 
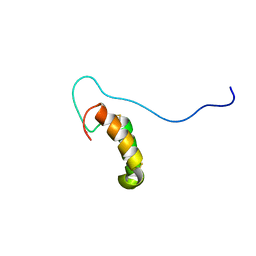 | | Solution structure of human Cu(I)Cox17 | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RN9
 
 | | Solution structure of human apoCox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
1AKK
 
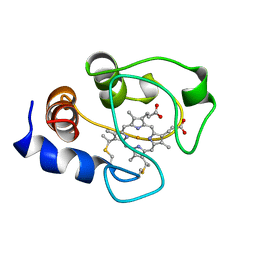 | | SOLUTION STRUCTURE OF OXIDIZED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Gray, H.B, Luchinat, C, Reddig, T, Rosato, A, Turano, P. | | Deposit date: | 1997-05-22 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized horse heart cytochrome c.
Biochemistry, 36, 1997
|
|
1FHB
 
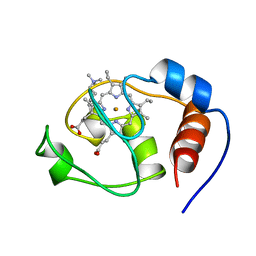 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE CYANIDE ADDUCT OF A MET80ALA VARIANT OF SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C. IDENTIFICATION OF LIGAND-RESIDUE INTERACTIONS IN THE DISTAL HEME CAVITY | | Descriptor: | CYANIDE ION, FERRICYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1995-06-16 | | Release date: | 1995-09-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Solution Structure of the Cyanide Adduct of a met80Ala Variant of Saccharomyces Cerevisiae Iso-1-Cytochrome C. Identification of Ligand-Residue Interactions in the Distal Heme Cavity
Biochemistry, 34, 1995
|
|
1FI7
 
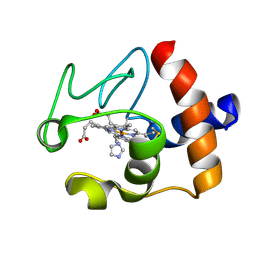 | | Solution structure of the imidazole complex of cytochrome C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FI9
 
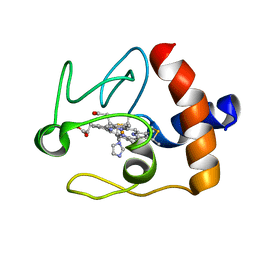 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1L3N
 
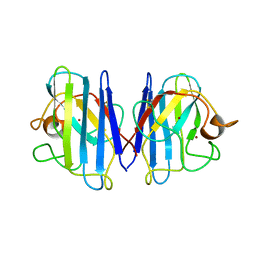 | | The Solution Structure of Reduced Dimeric Copper Zinc SOD: the Structural Effects of Dimerization | | Descriptor: | COPPER (I) ION, ZINC ION, superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-05-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization
Eur.J.Biochem., 269, 2002
|
|
1SO9
 
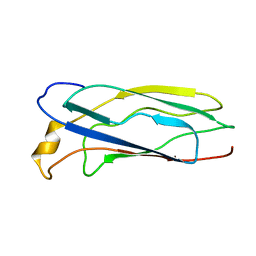 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1S4I
 
 | | Crystal structure of a SOD-like protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, ZINC ION, superoxide dismutase-like protein yojM | | Authors: | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | Deposit date: | 2004-01-16 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1AQA
 
 | | SOLUTION STRUCTURE OF REDUCED MICROSOMAL RAT CYTOCHROME B5, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Ferroni, F, Rosato, A. | | Deposit date: | 1997-07-28 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced microsomal rat cytochrome b5.
Eur.J.Biochem., 249, 1997
|
|
1JWW
 
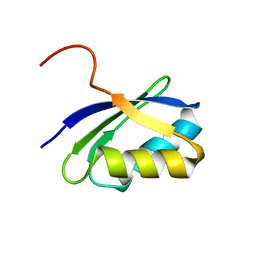 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
