1TU7
 
 | |
1OQS
 
 | | Crystal Structure of RV4/RV7 Complex | | Descriptor: | Phospholipase A2 RV-4, Phospholipase A2 RV-7 | | Authors: | Perbandt, M, Betzel, C. | | Deposit date: | 2003-03-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimeric neurotoxic complex viperotoxin F (RV-4/RV-7) from the venom of Vipera russelli formosensis at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2IEF
 
 | | Structure of the cooperative Excisionase (Xis)-DNA complex reveals a micronucleoprotein filament | | Descriptor: | 15-mer DNA, 19-mer DNA, 34-mer DNA, ... | | Authors: | Abbani, M.A, Papagiannis, C.V, Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HXA
 
 | |
2HX9
 
 | |
2HX7
 
 | |
2HX8
 
 | |
2F5V
 
 | | Reaction geometry and thermostability mutant of pyranose 2-oxidase from the white-rot fungus Peniophora sp. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Bastian, S, Heckmann-Pohl, D, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-28 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
2HNL
 
 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
1PA3
 
 | |
2F6C
 
 | | Reaction geometry and thermostability of pyranose 2-oxidase from the white-rot fungus Peniophora sp., Thermostability mutant E542K | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Heckmann-Pohl, D.M, Bastian, S, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-29 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
439D
 
 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1Q4J
 
 | |
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1JGC
 
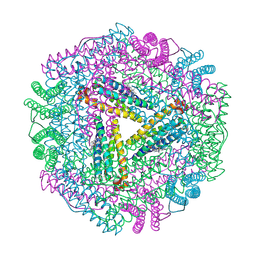 | | The 2.6 A Structure Resolution of Rhodobacter capsulatus Bacterioferritin with Metal-free Dinuclear Site and Heme Iron in a Crystallographic Special Position | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, bacterioferritin | | Authors: | Cobessi, D, Huang, L.-S, Ban, M, Pon, N.G, Daldal, F, Berry, E.A. | | Deposit date: | 2001-06-24 | | Release date: | 2002-01-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 A resolution structure of Rhodobacter capsulatus bacterioferritin with metal-free dinuclear site and heme iron in a crystallographic 'special position'.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A8X
 
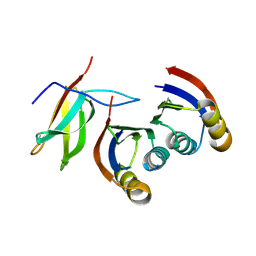 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with the HMA domain of Pikh-1 from rice (Oryza sativa) | | Descriptor: | AVR-Pik protein, NBS-LRR class disease resistance protein | | Authors: | Maidment, J.H.R, Xiao, G, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
4MEW
 
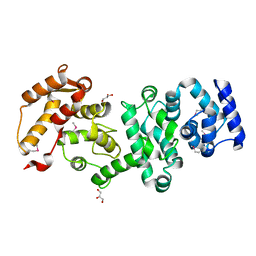 | | Structure of the core fragment of human PR70 | | Descriptor: | CALCIUM ION, GLYCEROL, Serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit beta | | Authors: | Dovega, R.B, Quistgaard, E.M, Tsutakawa, S, Anandapadamanaban, M, Low, C, Nordlund, P. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural and Biochemical Characterization of Human PR70 in Isolation and in Complex with the Scaffolding Subunit of Protein Phosphatase 2A.
Plos One, 9, 2014
|
|
4YGK
 
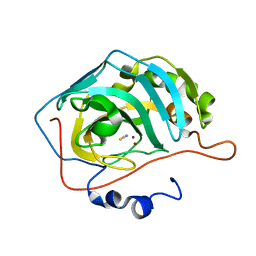 | | NaSCN--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, HYDROXIDE ION, THIOCYANATE ION, ... | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
7OWG
 
 | | human DEPTOR in a complex with mutant human mTORC1 A1459P | | Descriptor: | DEP domain-containing mTOR-interacting protein, Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, ... | | Authors: | Heimhalt, M, Berndt, A, Wagstaff, J, Anandapadamanaban, M, Perisic, O, Maslen, S, McLaughlin, S, Yu, W.-H, Masson, G.R, Boland, A, Ni, X, Yamashita, K, Murshudov, G.N, Skehel, M, Freund, S.M, Williams, R.L. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife, 10, 2021
|
|
3ZRG
 
 | | Crystal structure of RxLR effector PexRD2 from Phytophthora infestans | | Descriptor: | BROMIDE ION, PEXRD2 FAMILY SECRETED RXLR EFFECTOR PEPTIDE, PUTATIVE | | Authors: | King, S.R.F, Boutemy, L.S, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
4YDE
 
 | | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE | | Descriptor: | (3R,7S)-3,7,11-trimethyldodecyl trihydrogen diphosphate, 1,2-ETHANEDIOL, Protein farnesyltransferase/geranylgeranyltransferase type-1 Subunit beta, ... | | Authors: | Kumar, S, Mabanglo, M.F, Hast, M.A, Shi, Y, Beese, L.S. | | Deposit date: | 2015-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE
To Be Published
|
|
