3V4A
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 2 | | Descriptor: | (5R)-3-(3,4-dichlorophenyl)-5-(4-hydroxyphenyl)-1,5-dimethyl-2-thioxoimidazolidin-4-one, Androgen receptor, SULFATE ION | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
6QEY
 
 | | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties | | Descriptor: | ACETONITRILE, Insulin-like growth factor 2 mRNA-binding protein 1, PHOSPHATE ION | | Authors: | Dagil, R, Ball, N.J, Ogrodowicz, R.W, Purkiss, A.G, Taylor, I.A, Ramos, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties.
Nucleic Acids Res., 47, 2019
|
|
6FQF
 
 | | THE X-RAY STRUCTURE OF FERRIC-BETA4 HUMAN HEMOGLOBIN | | Descriptor: | Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Merlino, A, Balasco, N, Balsamo, A, Vergara, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ferric homotetrameric beta4human hemoglobin.
Biophys. Chem., 240, 2018
|
|
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
5MHP
 
 | | Novel Imidazo[1,2-a]pyridine Derivatives with Potent Autotaxin/ENPP2 Inhibitor Activity | | Descriptor: | 2-[[2-ethyl-8-methyl-6-[4-[2-(3-oxidanylazetidin-1-yl)-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fleury, D, Mueller, I, Lamers, M, Triballeau, N, Mollat, P, Vercheval, L. | | Deposit date: | 2016-11-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of 2-[[2-Ethyl-6-[4-[2-(3-hydroxyazetidin-1-yl)-2-oxoethyl]piperazin-1-yl]-8-methylimidazo[1,2-a]pyridin-3-yl]methylamino]-4-(4-fluorophenyl)thiazole-5-carbonitrile (GLPG1690), a First-in-Class Autotaxin Inhibitor Undergoing Clinical Evaluation for the Treatment of Idiopathic Pulmonary Fibrosis.
J. Med. Chem., 60, 2017
|
|
5M7M
 
 | | Novel Imidazo[1,2-a]pyridine Derivatives with Potent Autotaxin/ENPP2 Inhibitor Activity | | Descriptor: | CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, IODIDE ION, ... | | Authors: | Wolhkoning, A, Fleury, D, Leonard, P, Triballeau, N, Mollat, P, Vercheval, L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Structure-Activity Relationship, and Binding Mode of an Imidazo[1,2-a]pyridine Series of Autotaxin Inhibitors.
J. Med. Chem., 60, 2017
|
|
6Y16
 
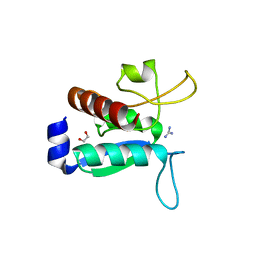 | | CRYSTAL STRUCTURE OF TMARGBP DOMAIN 1 IN COMPLEX WITH THE GUANIDINIUM ION | | Descriptor: | 1,2-ETHANEDIOL, Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, ... | | Authors: | Ruggiero, A, Balasco, N, Smaldone, G, Graziano, G, Vitagliano, L. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium binding to proteins: The intriguing effects on the D1 and D2 domains of Thermotoga maritima Arginine Binding Protein and a comprehensive analysis of the Protein Data Bank.
Int.J.Biol.Macromol., 163, 2020
|
|
6SAI
 
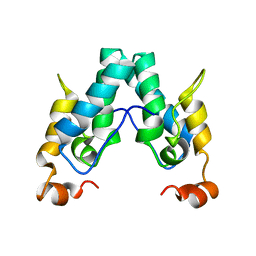 | |
6SA9
 
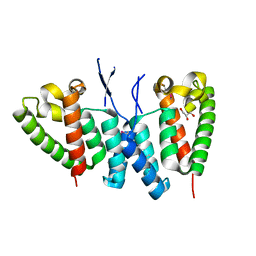 | |
6SVF
 
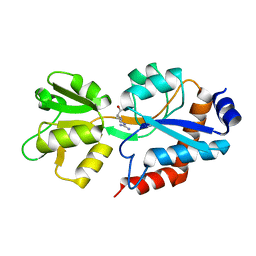 | | Crystal structure of the P235GK mutant of ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Vitagliano, L, Berisio, R, Esposito, L, Balasco, N, Smaldone, G, Ruggiero, A. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The non-swapped monomeric structure of the arginine-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5M1H
 
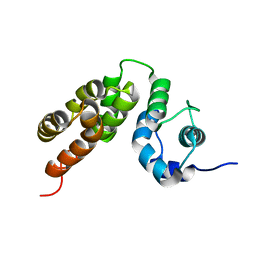 | |
6VWS
 
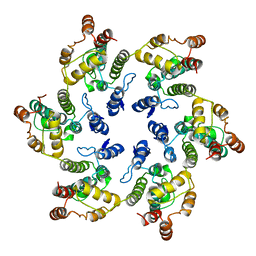 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
5M1G
 
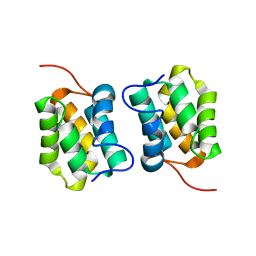 | |
8AO0
 
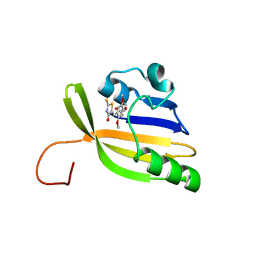 | | Solution structure of nanoFAST/HBR-DOM2 complex | | Descriptor: | (5~{Z})-5-[(2,5-dimethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Photoactive yellow protein | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baleeva, N.S, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
6ES4
 
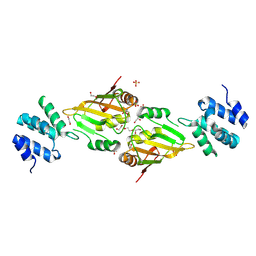 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
6GPD
 
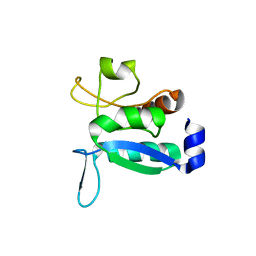 | | Crystal structure of the ligand-free form of domain 1 from TmArgBP | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
6GGP
 
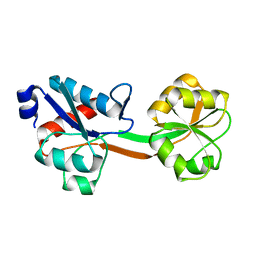 | | Structure of the ligand-free form of truncated ArgBP (residues 20-233) from T. maritima | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Berisio, R, Balasco, N, D'Auria, S, Vitagliano, L, Ruggiero, A. | | Deposit date: | 2018-05-03 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Domain swapping dissection in Thermotoga maritima arginine binding protein: How structural flexibility may compensate destabilization.
Biochim. Biophys. Acta, 1866, 2018
|
|
6GGV
 
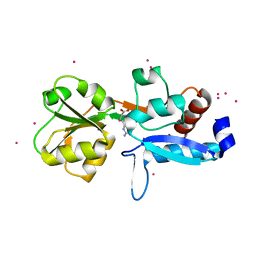 | | Structure of the arginine-bound form of truncated (residues 20-233) ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Smaldone, G, Berisio, R, Balasco, N, D'Auria, S, Vitagliano, L, Ruggiero, A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Domain swapping dissection in Thermotoga maritima arginine binding protein: How structural flexibility may compensate destabilization.
Biochim. Biophys. Acta, 1866, 2018
|
|
6AIE
 
 | | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution | | Descriptor: | Putative methyltransferase | | Authors: | Venkataraman, S, Dhankar, A, Sinha, K.M, Manivasakan, P, Iqbal, N, Singh, T.P, Prasad, B.V.L.S. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution
To Be Published
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
6K2M
 
 | |
5ZZV
 
 | | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution
To Be Published
|
|
5YIH
 
 | | Crystal structure of tetrameric Nucleoside diphosphate kinase at 1.98 A resolution from Acinetobacter baumannii | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Bairagya, H.R, Sikarwar, J, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of tetrameric Nucleoside diphosphate kinase at 1.98 A resolution from Acinetobacter baumannii
To Be Published
|
|
6J93
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase form apo at 0.95 A resolution.
To Be Published
|
|
5Y9A
 
 | | Crystal structure of the complex of peptidyl tRNA hydrolase with a phosphate ion at the substrate binding site and cytarabine at a new ligand binding site at 1.1 A resolution | | Descriptor: | CYTARABINE, PHOSPHATE ION, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Iqbal, N, Singh, N, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.
Biochem. J., 475, 2018
|
|
