6H8E
 
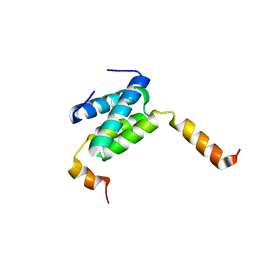 | | Truncated derivative of the C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
6HS5
 
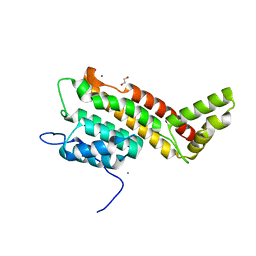 | | N-terminal domain including the conserved ImpA_N region of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TssA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-09-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
3MP9
 
 | | Structure of Streptococcal protein G B1 domain at pH 3.0 | | Descriptor: | FORMIC ACID, Immunoglobulin G-binding protein G | | Authors: | Tomlinson, J.H, Green, V.L, Baker, P.J, Williamson, M.P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural origins of pH-dependent chemical shifts in the B1 domain of protein G.
Proteins, 78, 2010
|
|
1K89
 
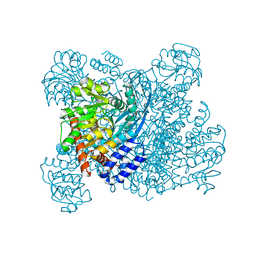 | | K89L MUTANT OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Stillman, T.J, Migueis, A.M.B, Wang, X.G, Baker, P.J, Britton, K.L, Engel, P.C, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the mechanism of domain closure and substrate specificity of glutamate dehydrogenase from Clostridium symbiosum.
J.Mol.Biol., 285, 1999
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
1X82
 
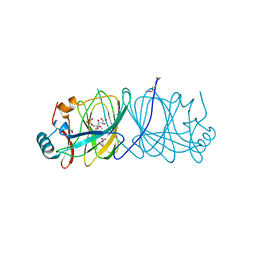 | | CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE FROM PYROCOCCUS FURIOSUS WITH BOUND 5-phospho-D-arabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X8E
 
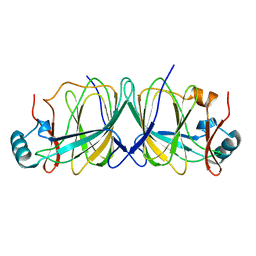 | | Crystal structure of Pyrococcus furiosus phosphoglucose isomerase free enzyme | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X7N
 
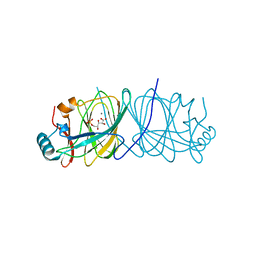 | | The crystal structure of Pyrococcus furiosus phosphoglucose isomerase with bound 5-phospho-D-arabinonate and Manganese | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, MANGANESE (II) ION | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-16 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
6ZZ5
 
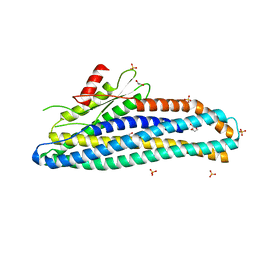 | | Structure of soluble SmhB of the tripartite alpha-pore forming toxin, Smh, from Serratia marcescens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SmhB | | Authors: | Churchill-Angus, A.M, Baker, P.J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Characterisation of a tripartite alpha-pore forming toxin from Serratia marcescens
Sci Rep, 11, 2021
|
|
6ZZH
 
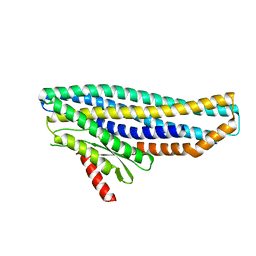 | |
7A27
 
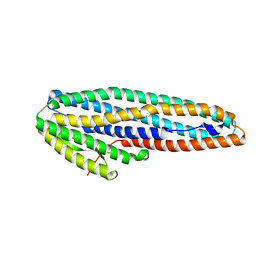 | |
7A0G
 
 | |
7A26
 
 | |
4MU3
 
 | | The form A structure of an E21Q catalytic mutant of A. thaliana IGPD2 in complex with Mn2+ and a mixture of its substrate, 2R3S-IGP, and an inhibitor, 2S3S-IGP, to 1.12 A resolution | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, (2S,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, 1,2-ETHANEDIOL, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4MU1
 
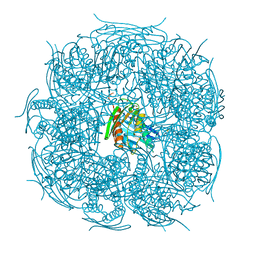 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+, imidazole, and sulfate at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4QNJ
 
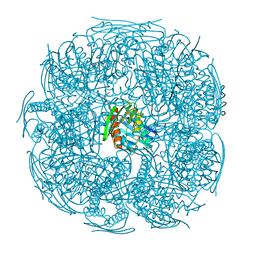 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and formate at 1.3A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4QNK
 
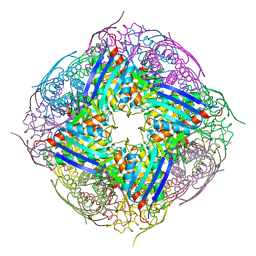 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
1Q8R
 
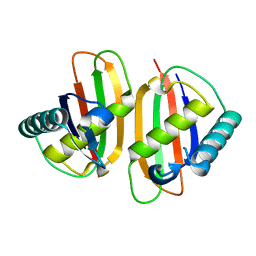 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
1SUL
 
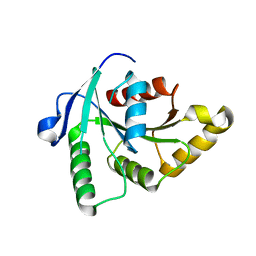 | | Crystal Structure of the apo-YsxC | | Descriptor: | GTP-binding protein YsxC | | Authors: | Ruzheinikov, S.N, Das, K.S, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SVI
 
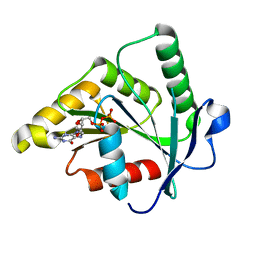 | | Crystal Structure of the GTP-binding protein YsxC complexed with GDP | | Descriptor: | GTP-binding protein YSXC, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
5EKW
 
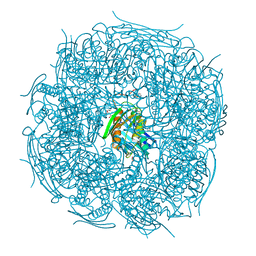 | | A. thaliana IGPD2 in complex with the racemate of the triazole-phosphonate inhibitor, C348 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5DNL
 
 | | Crystal structure of IGPD from Pyrococcus furiosus in complex with (S)-C348 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2S)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5DNX
 
 | | Crystal structure of IGPD from Pyrococcus furiosus in complex with (R)-C348 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5EL9
 
 | | A. thaliana IGPD2 in complex with the triazole-phosphonate inhibitor, (S)-C348, to 1.1A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5ELW
 
 | | A. thaliana IGPD2 in complex with the triazole-phosphonate inhibitor, (R)-C348, to 1.36A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
