4GVG
 
 | |
4GVF
 
 | |
4GVI
 
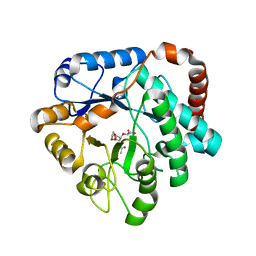 | | Crystal structure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc | | 分子名称: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bacik, J.P, Mark, B.L. | | 登録日 | 2012-08-30 | | 公開日 | 2012-12-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Active Site Plasticity within the Glycoside Hydrolase NagZ Underlies a Dynamic Mechanism of Substrate Distortion.
Chem.Biol., 19, 2012
|
|
4GYJ
 
 | |
4GVH
 
 | |
4GYK
 
 | |
3H8V
 
 | | Human Ubiquitin-activating Enzyme 5 in Complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | 著者 | Walker, J.R, Bacik, J.P, Rastgoo, N, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-04-29 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme.
J.Biol.Chem., 285, 2010
|
|
3GUC
 
 | | Human Ubiquitin-activating Enzyme 5 in Complex with AMPPNP | | 分子名称: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | 著者 | Walker, J.R, Bacik, J.P, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-03-29 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Human Ubiquitin-activating Enzyme 5 in Complex with AMPPNP
To be Published
|
|
5V1Q
 
 | |
5V1S
 
 | |
5V1T
 
 | | Crystal structure of Streptococcus suis SuiB bound to precursor peptide SuiA | | 分子名称: | IRON/SULFUR CLUSTER, METHIONINE, Radical SAM, ... | | 著者 | Davis, K.M, Bacik, J.P, Ando, N. | | 登録日 | 2017-03-02 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3PT2
 
 | | Structure of a viral OTU domain protease bound to Ubiquitin | | 分子名称: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | 著者 | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | 登録日 | 2010-12-02 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-05-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MT9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | 登録日 | 2018-10-19 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MV9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase, ... | | 著者 | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | 登録日 | 2018-10-24 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MYX
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | 著者 | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | 登録日 | 2018-11-02 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MW3
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | 著者 | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | 登録日 | 2018-10-29 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.65 Å) | | 主引用文献 | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MVE
 
 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | 登録日 | 2018-10-25 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
