5BMO
 
 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
5E43
 
 | | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum | | Descriptor: | ACETATE ION, Beta-lactamase, NITRATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-05 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7095 Å) | | Cite: | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum
To Be Published
|
|
3MKK
 
 | | The crystal structure of the D307A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with isomaltose | | Descriptor: | alpha-D-glucopyranose, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose, alpha-glucosidase GH31 family, ... | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structures Of The Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174
Faseb J., 24, 2010
|
|
3NSX
 
 | | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174
To be Published
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
3N04
 
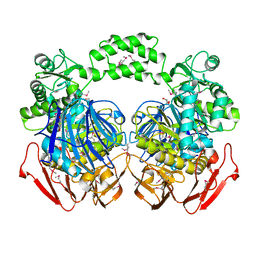 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | GLYCEROL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
4N1X
 
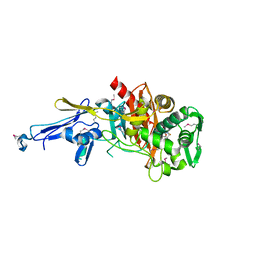 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G | | Descriptor: | OPEN FORM - PENICILLIN G, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G
To be Published
|
|
3POC
 
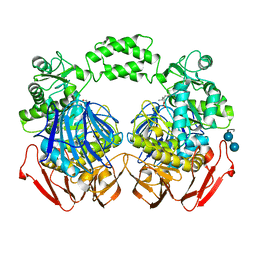 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
6NTR
 
 | | Crystal Structure of Beta-barrel-like Protein of Domain of Unknown Function DUF1849 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ATP/GTP-binding site-containing protein A, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-30 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
J.Bacteriol., 201, 2019
|
|
6NHS
 
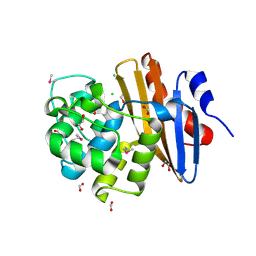 | | Crystal Structure of the Beta Lactamase Class D YbXI from Nostoc | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbXI from Nostoc
To Be Published
|
|
6NI0
 
 | | Crystal Structure of the Beta Lactamase Class D YbxI from Burkholderia thailandensis | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Kim, Y, Wu, R, Endres, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbxI from Burkholderia thailandensis
To Be Published
|
|
6OK0
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Sel1 repeat protein, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
4MQB
 
 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
6OK3
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6OZV
 
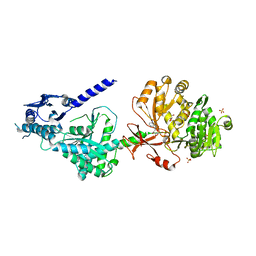 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-16 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6P1J
 
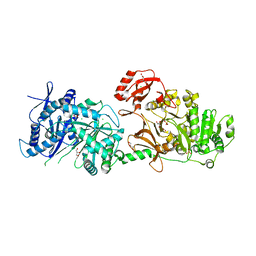 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo2 serine module | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-20 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
5C6U
 
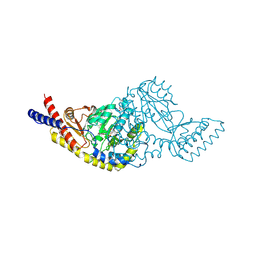 | | Rv3722c aminotransferase from Mycobacterium tuberculosis | | Descriptor: | Aminotransferase, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | OSIPIUK, J, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-15 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rv3722c aminotransferase from Mycobacterium tuberculosis.
to be published
|
|
6ONW
 
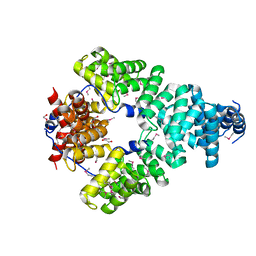 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6P4U
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with Mg and AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6OYF
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6P3I
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with Mg | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
4NEO
 
 | | Structure of BlmI, a type-II acyl-carrier-protein from Streptomyces verticillus involved in bleomycin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Peptide synthetase NRPS type II-PCP | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Babnigg, G, Bruno, C.J.P, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins.
Proteins, 82, 2014
|
|
4OPF
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NUK
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | ALPHA-GLUCOSIDASE, GLYCEROL | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174
TO BE PUBLISHED
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
