8E1I
 
 | | Asp1 kinase in complex with ATP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1S
 
 | | Asp1 kinase in complex with ADPNP Mn IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MANGANESE (II) ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1T
 
 | | Asp1 kinase in complex with ADPNP Mg IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1V
 
 | | Asp1 kinase in complex with ADPNP Mg IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1J
 
 | | Asp1 kinase in complex with 1,5-IP8 | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
1HHO
 
 | | STRUCTURE OF HUMAN OXYHAEMOGLOBIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN A (OXY) (ALPHA CHAIN), HEMOGLOBIN A (OXY) (BETA CHAIN), OXYGEN MOLECULE, ... | | Authors: | Shaanan, B. | | Deposit date: | 1983-06-10 | | Release date: | 1983-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human oxyhaemoglobin at 2.1 A resolution.
J.Mol.Biol., 171, 1983
|
|
4V2U
 
 | | Apo-structure of alpha2,3-sialyltransferase from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V0I
 
 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4W82
 
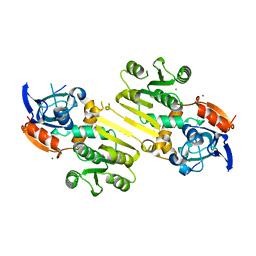 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase | | Descriptor: | CHLORIDE ION, Fatty acid synthase, MAGNESIUM ION, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
4V4Y
 
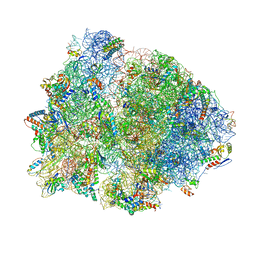 | | Crystal structure of the 70S Thermus thermophilus ribosome with translocated and rotated Shine-Dalgarno Duplex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4URE
 
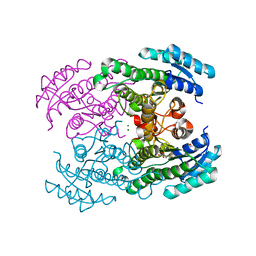 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
4UXA
 
 | | Improved variant of (R)-selective manganese-dependent hydroxynitrile lyase from bacteria | | Descriptor: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | Authors: | Pavkov-Keller, T, Wiedner, R, Kothbauer, B, Gruber-Khadjawi, M, Schwab, H, Steiner, K, Gruber, K. | | Deposit date: | 2014-08-21 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the Properties of Bacterial R-Selective Hydroxynitrile Lyases for Industrial Applications
Chemcatchem, 2015
|
|
1H96
 
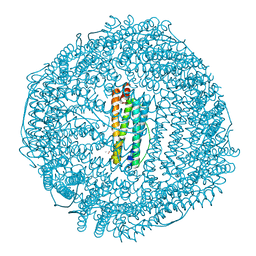 | | recombinant mouse L-chain ferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, SULFATE ION | | Authors: | Granier, T, Gallois, B, D'Estaintot, B.L, Dautant, A, Chevalier, J.M, Mellado, J.M, Beaumont, C, Santambrogio, P, Arosio, P, Precigoux, G. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mouse L-Chain Ferritin at 1.6 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4V4D
 
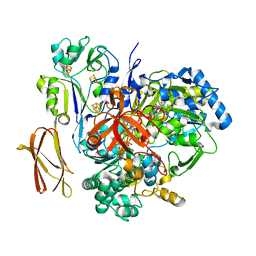 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with pyrogallol | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4V4Z
 
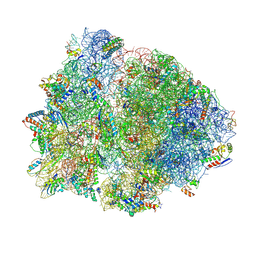 | | 70S Thermus thermophilous ribosome functional complex with mRNA and E- and P-site tRNAs at 4.5A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4URD
 
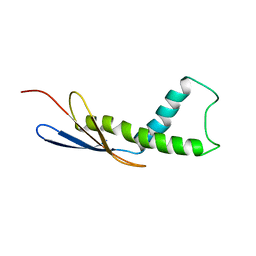 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
4UTP
 
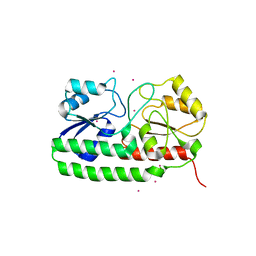 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4V5V
 
 | | Structure of respiratory syncytial virus nucleocapsid protein, P1 crystal form | | Descriptor: | RESPIRATORY SYNCYTIAL VIRUS NUCLEOCAPSID PROTEIN, RNA | | Authors: | El Omari, K, Dhaliwal, B, Ren, J, Abrescia, N.G.A, Lockyer, M, Powell, K.L, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-05-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of Respiratory Syncytial Virus Nucleocapsid Protein from Two Crystal Forms: Details of Potential Packing Interactions in the Native Helical Form.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4W7I
 
 | | Crystal structure of DEH reductase A1-R' mutant | | Descriptor: | 4-deoxy-L-erythro-5-hexoseulose uronate reductase A1-R' | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4W7S
 
 | | Crystal structure of the yeast DEAD-box splicing factor Prp28 at 2.54 Angstroms resolution | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jacewicz, A, Smith, P, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Nucleic Acids Res., 42, 2014
|
|
6L2Q
 
 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2019-10-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel tRNA-amino acid dual-site inhibitors against threonyl-tRNA synthetase by fragment-based target hopping.
Eur.J.Med.Chem., 187, 2019
|
|
4UTO
 
 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4V66
 
 | | Structure of the E. coli ribosome and the tRNAs in Post-accommodation state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Devkota, B, Caulfield, T.R, Tan, R.-Z, Harvey, S.C. | | Deposit date: | 2008-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Structure of the E. coli Ribosome Before and After Accommodation: Implications for Proofreading
To be Published
|
|
1HOY
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN A-BUNGAROTOXIN AND A MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Niccolai, N, Neri, P. | | Deposit date: | 2000-12-12 | | Release date: | 2000-12-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
6L0L
 
 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
