4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | 分子名称: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | 著者 | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | 登録日 | 2014-10-15 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4UU9
 
 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | 分子名称: | COMPLEMENT C5, MEDI7814, SULFATE ION | | 著者 | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | 登録日 | 2014-07-25 | | 公開日 | 2015-08-12 | | 最終更新日 | 2019-02-27 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2014-07-09 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4URN
 
 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | 分子名称: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-07-01 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4W2O
 
 | |
4V53
 
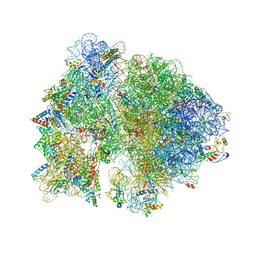 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin. | | 分子名称: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | 著者 | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | 登録日 | 2007-06-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.54 Å) | | 主引用文献 | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4W2P
 
 | |
4W2Q
 
 | |
4V5W
 
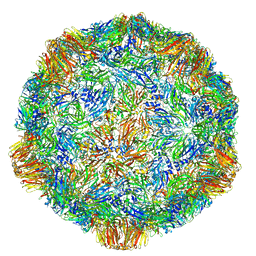 | | Grapevine Fanleaf virus | | 分子名称: | COAT PROTEIN | | 著者 | Schellenberger, P, Demangeat, G, Ritzenthaler, C, Lorber, B, Sauter, C. | | 登録日 | 2011-05-10 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
1VWK
 
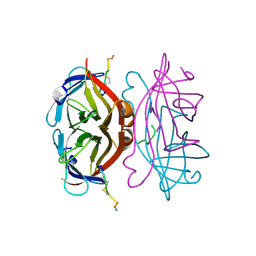 | | STREPTAVIDIN-CYCLO-[5-S-VALERAMIDE-HPQGPPC]K-NH2 | | 分子名称: | PENTANOIC ACID, PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | 著者 | Katz, B.A, Cass, R.T. | | 登録日 | 1997-03-03 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWA
 
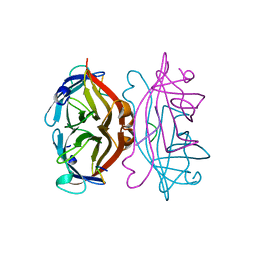 | | STREPTAVIDIN-FSHPQNT | | 分子名称: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | 著者 | Katz, B.A, Cass, R.T. | | 登録日 | 1997-03-03 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWI
 
 | |
1W1T
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(His-L-Pro) at 1.9 A resolution | | 分子名称: | CHITINASE B, CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | 著者 | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, van Aalten, D.M.F. | | 登録日 | 2004-06-24 | | 公開日 | 2005-01-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
1VWJ
 
 | |
1EO5
 
 | |
1W5C
 
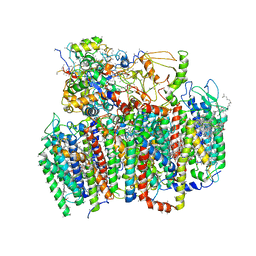 | | Photosystem II from Thermosynechococcus elongatus | | 分子名称: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | 著者 | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | 登録日 | 2004-08-06 | | 公開日 | 2004-12-21 | | 最終更新日 | 2014-01-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
6XN0
 
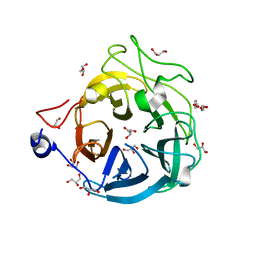 | | Crystal structure of GH43_1 enzyme from Xanthomonas citri | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | 登録日 | 2020-07-02 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.709 Å) | | 主引用文献 | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
1W0W
 
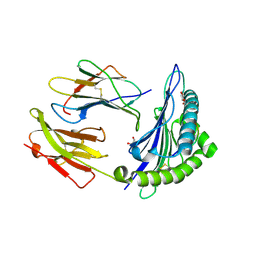 | | Crystal Structure Of HLA-B*2709 Complexed With the self-Peptide TIS from EGF-response factor 1 | | 分子名称: | BETA-2-MICROGLOBULIN, BUTYRATE RESPONSE FACTOR 2, GLYCEROL, ... | | 著者 | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | 登録日 | 2004-06-14 | | 公開日 | 2005-03-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Thermodynamic and Structural Equivalence of Two Hla-B27 Subtypes Complexed with a Self-Peptide
J.Mol.Biol., 346, 2005
|
|
6XB4
 
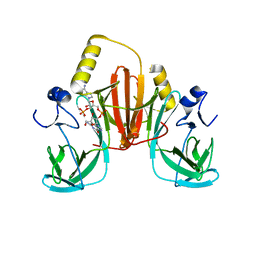 | |
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2021-02-05 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6XUC
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | 分子名称: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | 著者 | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | 登録日 | 2020-01-17 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8702 Å) | | 主引用文献 | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
1ED4
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH IPITU (H4B FREE) | | 分子名称: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | 著者 | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | 登録日 | 2000-01-26 | | 公開日 | 2000-10-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
7NHQ
 
 | | Structure of PSII-I prime (PSII with Psb28, and Psb34) | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | 著者 | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | 登録日 | 2021-02-11 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
1E97
 
 | |
1EA2
 
 | | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Tyrosine-to-Phenylalanine Substitution(s) in the Hydrogen Bond Network of Delta-5-3-Ketosteroid Isomerase from Pheudomonas putida Biotype B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Choi, G, Ha, N.-C, Kim, M.-S, Hong, B.-H, Choi, K.Y. | | 登録日 | 2000-11-03 | | 公開日 | 2001-11-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Substitution(S) of Tyrosine with Phenylalanine in the Hydrogen Bond Network of Delta (5)-3-Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochemistry, 40, 2001
|
|
