3VHE
 
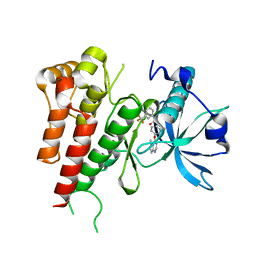 | | Crystal structure of human VEGFR2 kinase domain with a novel pyrrolopyrimidine inhibitor. | | Descriptor: | 1-{2-fluoro-4-[(5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Vascular endothelial growth factor receptor 2 | | Authors: | Oguro, Y, Miyamoto, N, Okada, K, Takagi, T, Iwata, H, Awazu, Y, Miki, H, Hori, A, Kamiyama, K, Imanura, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, synthesis, and evaluation of 5-methyl-4-phenoxy-5H-pyrrolo[3,2-d]pyrimidine derivatives: novel VEGFR2 kinase inhibitors binding to inactive kinase conformation.
Bioorg.Med.Chem., 18, 2010
|
|
2ZJB
 
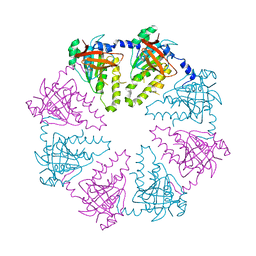 | | Crystal structure of the human Dmc1-M200V polymorphic variant | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Hikiba, J, Hirota, K, Kagawa, W, Ikawa, S, Kinebuchi, T, Sakane, I, Takizawa, Y, Yokoyama, S, Mandon-Pepin, B, Nicolas, A, Shibata, T, Ohta, K, Kurumizaka, H. | | Deposit date: | 2008-03-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional analyses of the DMC1-M200V polymorphism found in the human population
Nucleic Acids Res., 36, 2008
|
|
1DIZ
 
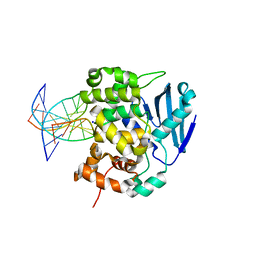 | | CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, DNA (5'-D(*GP*AP*CP*AP*TP*GP*AP*(NRI)P*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Hollis, T, Ichikawa, Y, Ellenberger, T.E. | | Deposit date: | 1999-11-30 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA.
EMBO J., 19, 2000
|
|
1IY3
 
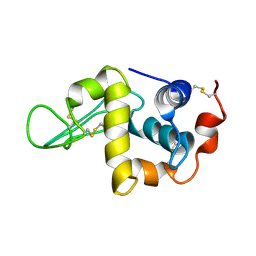 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IY4
 
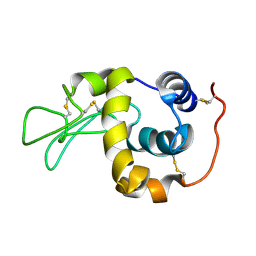 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1J32
 
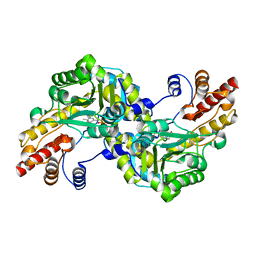 | |
2RPV
 
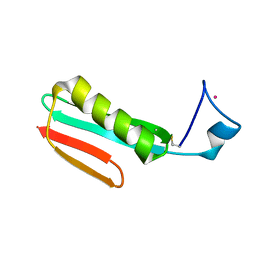 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
3W2S
 
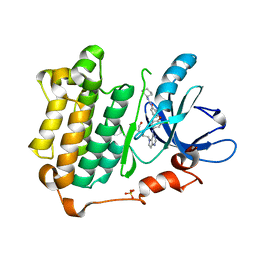 | | EGFR kinase domain with compound4 | | Descriptor: | 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W32
 
 | | EGFR kinase domain complexed with compound 20a | | Descriptor: | 4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-N-[2-(methylsulfonyl)ethyl]-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
3W2O
 
 | | EGFR Kinase domain T790M/L858R Mutant with TAK-285 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-3-hydroxy-3-methylbutanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
1JFL
 
 | | CRYSTAL STRUCTURE DETERMINATION OF ASPARTATE RACEMASE FROM AN ARCHAEA | | Descriptor: | ASPARTATE RACEMASE | | Authors: | Liu, L.J, Iwata, K, Kita, A, Kawarabayasi, Y, Yohda, M, Miki, K. | | Deposit date: | 2001-06-21 | | Release date: | 2002-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartate racemase from Pyrococcus horikoshii OT3 and its implications for molecular mechanism of PLP-independent racemization.
J.Mol.Biol., 319, 2002
|
|
1LMR
 
 | | Solution of ADO1, a Toxin from the Assassin Bugs Agriosphodrus dohrni that Blocks the Voltage Sensitive Calcium Channel L-type | | Descriptor: | TOXIN ADO1 | | Authors: | Bernard, C, Corzo, G, Adachi-Akahane, S, Foures, G, Kanemaru, K, Furukawa, Y, Nakajima, T, Darbon, H. | | Deposit date: | 2002-05-02 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ADO1, a toxin extracted from the saliva of the assassin bug, Agriosphodrus dohrni
Proteins: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3W2P
 
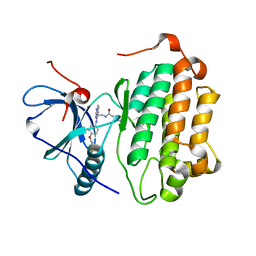 | | EGFR Kinase domain T790M/L858R mutant with compound 2 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-4-(dimethylamino)butanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2R
 
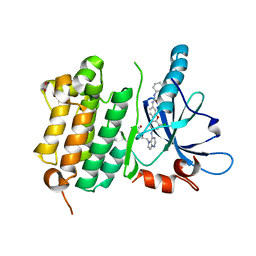 | | EGFR Kinase domain T790M/L858R mutant with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
2KE4
 
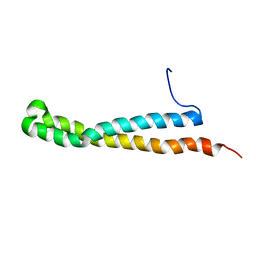 | |
3W2Q
 
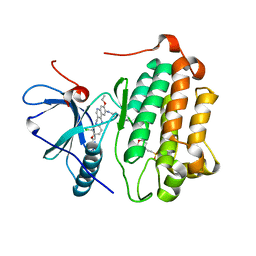 | | EGFR kinase domain T790M/L858R mutant with HKI-272 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W33
 
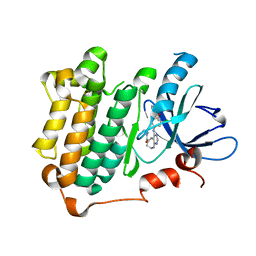 | | EGFR kinase domain complexed with compound 19b | | Descriptor: | 4-{[4-(1-benzothiophen-4-yloxy)-3-chlorophenyl]amino}-N-(2-hydroxyethyl)-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
2KBT
 
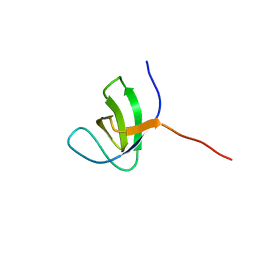 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | Descriptor: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | Authors: | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2008-12-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
4BGW
 
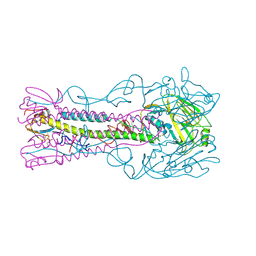 | | Crystal Structure of H5 (VN1194) Influenza Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
4BH4
 
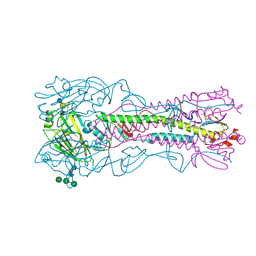 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH3
 
 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Human Receptor Analogue 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BGY
 
 | | H5 (VN1194) Influenza Virus Haemagglutinin in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
3VGV
 
 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
2ZZD
 
 | | Recombinant thiocyanate hydrolase, air-oxidized form of holo-enzyme | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
3VGT
 
 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
