8JH3
 
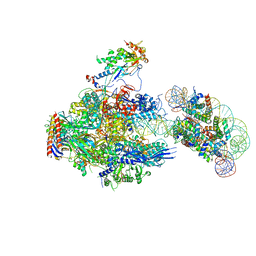 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2023-05-22 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
4Z5T
 
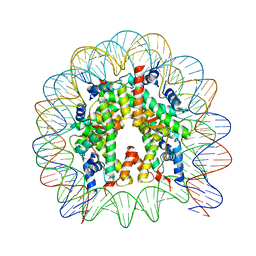 | | The nucleosome containing human H3.5 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | 登録日 | 2015-04-03 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
2YRN
 
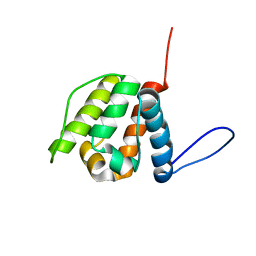 | | Solution structure of the CH domain from Human Neuron navigator 2 | | 分子名称: | Neuron navigator 2 isoform 4 | | 著者 | Tomizawa, T, Tochio, N, Koshiba, S, Inoue, M, Nakamura, Y, Furukawa, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-02 | | 公開日 | 2008-02-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the CH domain from Human Neuron navigator 2
To be Published
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
3J6P
 
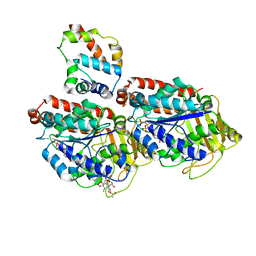 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | 分子名称: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | 登録日 | 2014-03-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
8JH2
 
 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | 著者 | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2023-05-22 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
3VX6
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | 分子名称: | E1 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8J91
 
 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones | | 分子名称: | DNA (169-MER), HTA13, Histone H2B.6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1
Nat Commun, 2024
|
|
8J90
 
 | | Cryo-EM structure of DDM1-nucleosome complex | | 分子名称: | ATP-dependent DNA helicase DDM1, DNA (169-MER), HTA6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.71 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1
Nat Commun, 2024
|
|
8J92
 
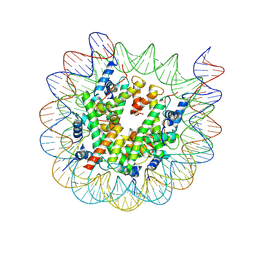 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W | | 分子名称: | DNA (169-MER), HTA6, HTB9, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1
Nat Commun, 2024
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | 分子名称: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2023-08-08 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
3VX7
 
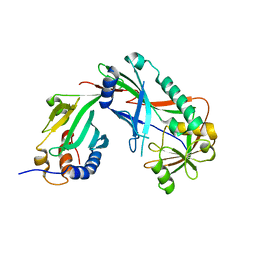 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | 分子名称: | E1, E2 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5AON
 
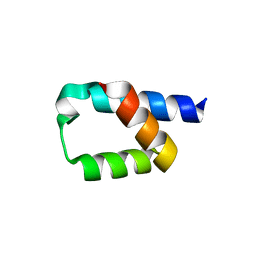 | | Crystal structure of the conserved N-terminal domain of Pex14 from Trypanosoma brucei | | 分子名称: | PEROXIN 14, SULFATE ION | | 著者 | Obita, T, Sugawara, Y, Mizuguchi, M, Watanabe, Y, Kawaguchi, K, Imanaka, T. | | 登録日 | 2015-09-11 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.646 Å) | | 主引用文献 | Characterization of the Interaction between Trypanosoma Brucei Pex5P and its Receptor Pex14P.
FEBS Lett., 590, 2016
|
|
6CX1
 
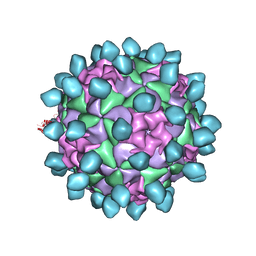 | | Cryo-EM structure of Seneca Valley Virus-Anthrax Toxin Receptor 1 complex | | 分子名称: | Anthrax toxin receptor 1, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Jayawardena, N, Burga, L, Easingwood, R, Takizawa, Y, Wolf, M, Bostina, M. | | 登録日 | 2018-04-02 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for anthrax toxin receptor 1 recognition by Seneca Valley Virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5B36
 
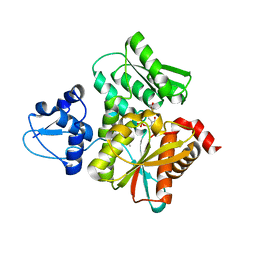 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | 登録日 | 2016-02-10 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
2XXP
 
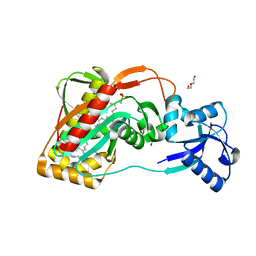 | | A widespread family of bacterial cell wall assembly proteins | | 分子名称: | CPS2A, DI(HYDROXYETHYL)ETHER, MONO-TRANS, ... | | 著者 | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | 登録日 | 2010-11-11 | | 公開日 | 2011-10-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
8PXL
 
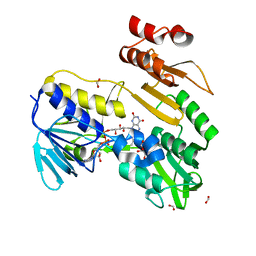 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 1.37 A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXK
 
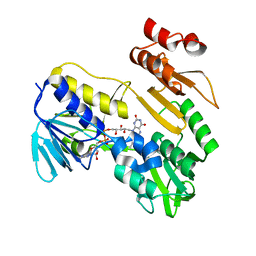 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 5.76 A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.77 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
2XXQ
 
 | | A widespread family of bacterial cell wall assembly proteins | | 分子名称: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | 登録日 | 2010-11-11 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
5B3A
 
 | | Crystal Structure of O-Phoshoserine Sulfhydrylase from Aeropyrum pernix in Complexed with the alpha-Aminoacrylate Intermediate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Protein CysO | | 著者 | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | 登録日 | 2016-02-12 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
