5YXM
 
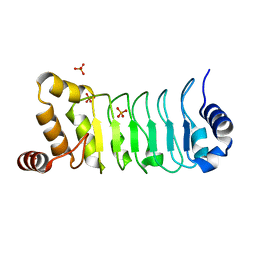 | | Crystal structure of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Dynein light chain 1, axonemal, PHOSPHATE ION | | Authors: | Toda, A, Tanaka, H, Nishikawa, Y, Yagi, T, Kurisu, G. | | Deposit date: | 2017-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structural atlas of dynein motors at atomic resolution.
Biophys Rev, 10, 2018
|
|
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
3D7A
 
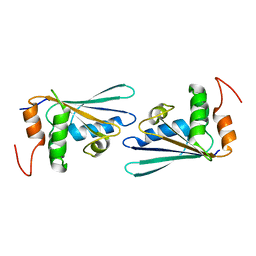 | |
1WQO
 
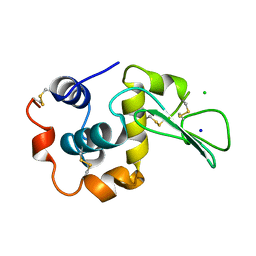 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQP
 
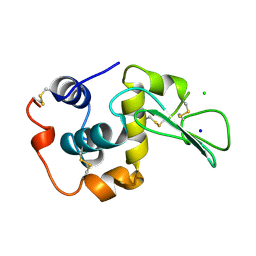 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
2KFJ
 
 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | Deposit date: | 2009-02-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
1SKY
 
 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
7BUW
 
 | | Eucommia ulmoides TPT3 mutant -C94Y/A95F | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUU
 
 | | Eucommia ulmoides TPT3, crystal form 1 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
3VX6
 
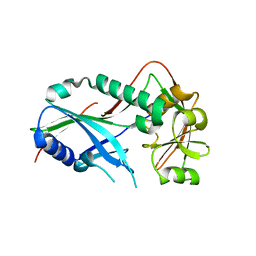 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7CBX
 
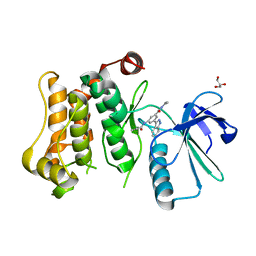 | |
3AP3
 
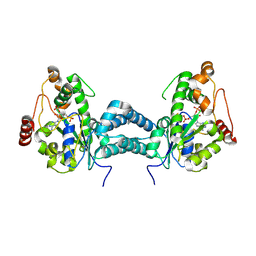 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Protein-tyrosine sulfotransferase 2 | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
3AP1
 
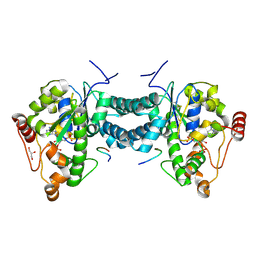 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
5H0X
 
 | |
2EK7
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M) | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M)
To be Published
|
|
1WQM
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQN
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQQ
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQR
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
2D2A
 
 | | Crystal Structure of Escherichia coli SufA Involved in Biosynthesis of Iron-sulfur Clusters | | Descriptor: | SufA protein | | Authors: | Wada, K, Hasegawa, Y, Gong, Z, Minami, Y, Fukuyama, K, Takahashi, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer
Febs Lett., 579, 2005
|
|
2D3W
 
 | | Crystal Structure of Escherichia coli SufC, an ATPase compenent of the SUF iron-sulfur cluster assembly machinery | | Descriptor: | Probable ATP-dependent transporter sufC | | Authors: | Kitaoka, S, Wada, K, Hasegawa, Y, Minami, Y, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery
Febs Lett., 580, 2006
|
|
3AP2
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP,C4 peptide, and phosphate ion | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2: Insights into substrate-binding and catalysis of post-translational protein tyrosine sulfation
To be Published
|
|
2DKV
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | Authors: | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
3KDC
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10074 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dichlorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3KDD
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10265 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-difluorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
