8TXA
 
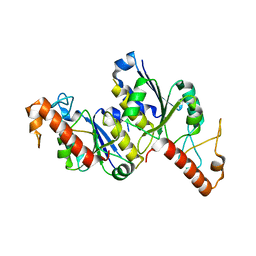 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
4JBI
 
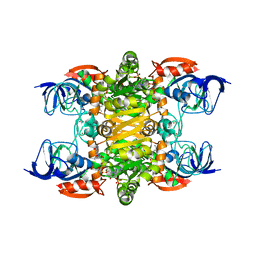 | | 2.35A resolution structure of NADPH bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
4TK5
 
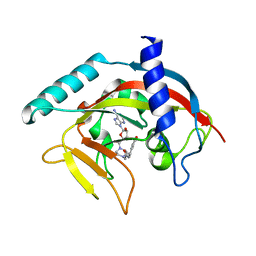 | | Crystal Structure of human Tankyrase 2 in complex with EB47. | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TJY
 
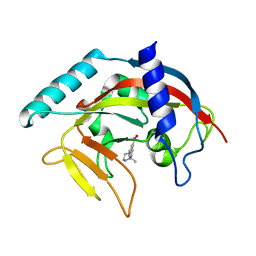 | | Crystal Structure of human Tankyrase 2 in complex with ABT-888. | | Descriptor: | 2-[(2S)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-7-carboxamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKI
 
 | | Crystal Structure of human Tankyrase 2 in complex with BSI-201. | | Descriptor: | 4-iodo-3-nitrobenzamide, ISOPROPYL ALCOHOL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7K7E
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 7.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7K7D
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 6.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
4TKF
 
 | | Crystal Structure of human Tankyrase 2 in complex with IWR-1. | | Descriptor: | 3-aminobenzamide, 4-[(3aR,4R,7S,7aS)-1,3-dioxooctahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TJU
 
 | | Crystal Structure of human Tankyrase 2 in complex with 3,4-CPQ-5-C. | | Descriptor: | 3-(4-CHLOROPHENYL)QUINOXALINE-5-CARBOXAMIDE, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7K7C
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.5 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
4TK0
 
 | | Crystal Structure of human Tankyrase 2 in complex with DPQ. | | Descriptor: | 5-[4-(piperidin-1-yl)butoxy]-3,4-dihydroisoquinolin-1(2H)-one, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7K7B
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
3Q8C
 
 | | Crystal structure of Protective Antigen W346F (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8A
 
 | | Crystal structure of WT Protective Antigen (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Rajapaksha, M, Lovell, S, Janowiak, B.E, Andra, K.K, Battaile, K.P, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8B
 
 | | Crystal structure of WT Protective Antigen (pH 9.0) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8E
 
 | | Crystal structure of Protective Antigen W346F (pH 8.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8F
 
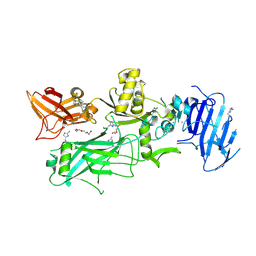 | | Crystal structure of 2-Fluorohistine labeled Protective Antigen (pH 5.8) | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3R9I
 
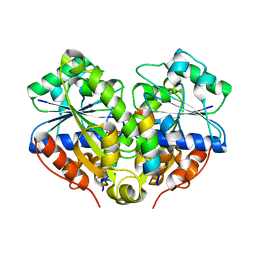 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
4LQU
 
 | | 1.60A resolution crystal structure of a superfolder green fluorescent protein (W57G) mutant | | Descriptor: | Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
3P8K
 
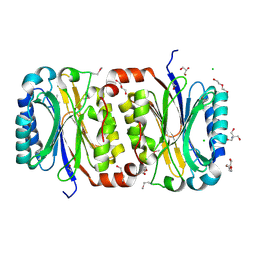 | | Crystal Structure of a putative carbon-nitrogen family hydrolase from Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gordon, R.D, Qiu, W, Battaile, K, Lam, K, Soloveychik, M, Benetteraj, D, Romanov, V, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-10-14 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of carbon-nitrogen family hydrolase from Staphylococcus aureus
To be Published
|
|
8W1D
 
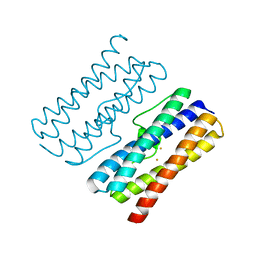 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
3P8A
 
 | | Crystal Structure of a hypothetical protein from Staphylococcus aureus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lam, R, Qiu, W, Battaile, K, Lam, K, Romanov, V, Chan, T, Pai, E, Chirgadze, N.Y. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a hypothetical protein from Staphylococcus aureus
To be Published
|
|
4JBH
 
 | | 2.2A resolution structure of cobalt and zinc bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
4LQT
 
 | | 1.10A resolution crystal structure of a superfolder green fluorescent protein (W57A) mutant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
