6QX0
 
 | | HEWL lysozyme, crystallized from LiCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6RRO
 
 | |
6RRL
 
 | |
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
6MLU
 
 | | Cryo-EM structure of lipid droplet formation protein Seipin/BSCL2 | | Descriptor: | Seipin | | Authors: | Sui, X, Arlt, H, Liao, M, Walther, C.T, Farese, V.R. | | Deposit date: | 2018-09-28 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of the lipid droplet-formation protein seipin.
J. Cell Biol., 217, 2018
|
|
6T6K
 
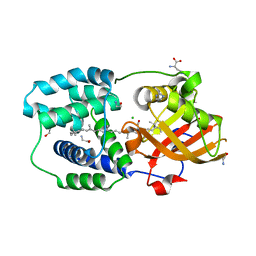 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCINE, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T6M
 
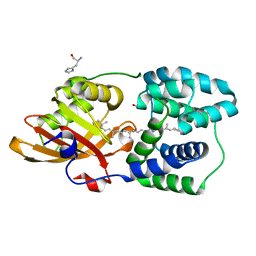 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | Descriptor: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T6O
 
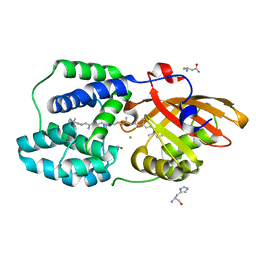 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 4.6 | | Descriptor: | ASPARAGINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
4L38
 
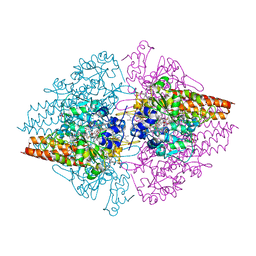 | | Nitrite complex of TvNiR, low dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-05 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
4L3Y
 
 | | Nitrite complex of TvNiR, high dose data set (NO complex) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
8QUA
 
 | | GTP binding protein YsxC from Staphylococcus aureus | | Descriptor: | ACETYL GROUP, GLYCEROL, Probable GTP-binding protein EngB | | Authors: | Biktimirov, A, Islamov, D, Lazarenko, V, Fatkhullin, B, Validov, S, Yusupov, M, Usachev, K. | | Deposit date: | 2023-10-15 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GTPase YsxC from Staphylococcus aureus.
Biochem.Biophys.Res.Commun., 699, 2024
|
|
4L3X
 
 | | Nitrite complex of TvNiR, first middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
4L3Z
 
 | | Nitrite complex of TvNiR, second middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
3QAY
 
 | | Catalytic domain of CD27L endolysin targeting Clostridia Difficile | | Descriptor: | Endolysin, PHOSPHATE ION, ZINC ION | | Authors: | Mayer, M.J, Garefaliki, V, Spoerl, R, Narbad, A, Meijers, R. | | Deposit date: | 2011-01-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range.
J.Bacteriol., 193, 2011
|
|
5EW1
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5EW2
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
4CU5
 
 | | C-terminal domain of endolysin from phage CD27L is a trigger and release factor | | Descriptor: | ENDOLYSIN | | Authors: | Dunne, M, Mertens, H.D.T, Garefalaki, V, Jeffries, C.M, Thompson, A, Lemke, E.A, Svergun, D.I, Mayer, M.J, Narbad, A, Meijers, R. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Cd27L and Ctp1L Endolysins Targeting Clostridia Contain a Built-in Trigger and Release Factor.
Plos Pathog., 10, 2014
|
|
2JST
 
 | | Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets II: Halothane Effects on Structure and Dynamics | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, Four-Alpha-Helix Bundle | | Authors: | Cui, T, Bondarenko, V, Ma, D, Canlas, C, Brandon, N.R, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2007-07-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part II: halothane effects on structure and dynamics
Biophys.J., 94, 2008
|
|
2H3K
 
 | | Solution Structure of the first NEAT domain of IsdH | | Descriptor: | Haptoglobin-binding surface anchored protein | | Authors: | Pilpa, R.M, Fadeev, E.A, Villareal, V.A, Wong, M.A, Phillips, M, Clubb, R.T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus.
J.Mol.Biol., 360, 2006
|
|
2KID
 
 | | Solution Structure of the S. Aureus Sortase A-substrate Complex | | Descriptor: | (PHQ)LPA(B27) peptide, CALCIUM ION, Sortase | | Authors: | Suree, N, Liew, C.K, Villareal, V.A, Thieu, W, Fadeev, E.A, Clemens, J.J, Jung, M.E, Clubb, R.T. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized.
J.Biol.Chem., 284, 2009
|
|
4CU2
 
 | | C-terminal domain of CTP1L endolysin mutant V195P that reduces autoproteolysis | | Descriptor: | ENDOLYSIN | | Authors: | Dunne, M, Mertens, H.D.T, Garefalaki, V, Jeffries, C.M, Thompson, A, Lemke, E.A, Svergun, D.I, Mayer, M.J, Narbad, A, Meijers, R. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Cd27L and Ctp1L Endolysins Targeting Clostridia Contain a Built-in Trigger and Release Factor.
Plos Pathog., 10, 2014
|
|
2I7U
 
 | | Structural and Dynamical Analysis of a Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets | | Descriptor: | Four-alpha-helix bundle | | Authors: | Ma, D, Brandon, N.R, Cui, T, Bondarenko, V, Canlas, C, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part I: structural and dynamical analyses.
Biophys.J., 94, 2008
|
|
2M61
 
 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
