5XEV
 
 | | Crystal Structure of a novel Xaa-Pro dipeptidase from Deinococcus radiodurans | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Are, V.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a novel prolidase from Deinococcus radiodurans identifies new subfamily of bacterial prolidases.
Proteins, 85, 2017
|
|
5GIQ
 
 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | Authors: | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
5GIU
 
 | | Crystal structure of Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, SODIUM ION | | Authors: | Are, V.N, Kumar, A, Singh, R, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site
To Be Published
|
|
5CE6
 
 | | N-terminal domain of FACT complex subunit SPT16 from Cicer arietinum (chickpea) | | Descriptor: | ACETATE ION, FACT-Spt16, POTASSIUM ION, ... | | Authors: | Are, V.N, Ghosh, B, Kumar, A, Makde, R. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum.
Int.J.Biol.Macromol., 88, 2016
|
|
7P1Z
 
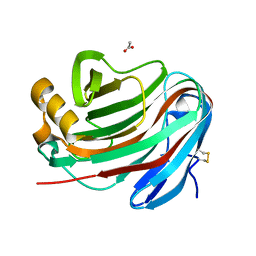 | | Novel GH12 endogluconase from Aspergillus cervinus | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Lazarenko, V.A, Rykov, S.V, Nikolaeva, A.Y, Berezina, O.V, Akentyev, F.I. | | Deposit date: | 2021-07-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Unusual substrate specificity in GH family 12: structure-function analysis of glucanases Bgh12A and Xgh12B from Aspergillus cervinus, and Egh12 from Thielavia terrestris.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
1XLT
 
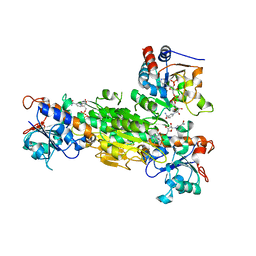 | | Crystal structure of Transhydrogenase [(domain I)2:domain III] heterotrimer complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sundaresan, V, Chartron, J, Yamaguchi, M, Stout, C.D. | | Deposit date: | 2004-09-30 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational Diversity in NAD(H) and Transhydrogenase Nicotinamide Nucleotide Binding Domains
J.Mol.Biol., 346, 2005
|
|
6D90
 
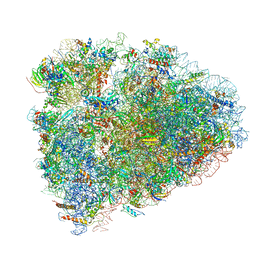 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-site tRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-27 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
6D9J
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-sitetRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
8F4V
 
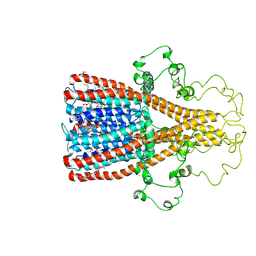 | | Alpha7 nicotinic acetylcholine receptor intracellular and transmembrane domains bound to ivermectin in a desensitized state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Neuronal acetylcholine receptor subunit alpha-7 | | Authors: | Bondarenko, V, Chen, Q, Tang, P. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of Ivermectin Binding to alpha 7nAChR and the Induced Channel Desensitization.
Acs Chem Neurosci, 14, 2023
|
|
4YMZ
 
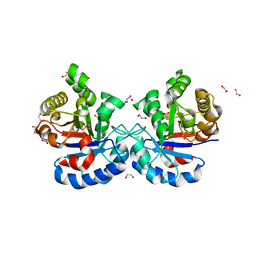 | | DHAP bound Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, SULFATE ION, ... | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-08 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Z0J
 
 | | F96S/S73A Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Z0S
 
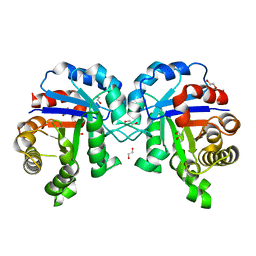 | |
4YXG
 
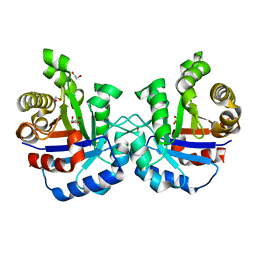 | |
7RPM
 
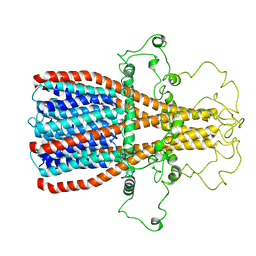 | |
4X22
 
 | | Crystal structure of Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4YWI
 
 | | F96S/L167V Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-20 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Q4U
 
 | | TvNiR in complex with sulfite, low dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
to be published
|
|
4Q17
 
 | | Free form of TvNiR, middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-03 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be published
|
|
4Q0T
 
 | | Free form of TvNiR, low dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-02 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
TO BE PUBLISHED
|
|
4Q5B
 
 | | TvNiR in complex with sulfite, high dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be Published
|
|
4Q1O
 
 | | Free form of TvNiR, high dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-04 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be Published
|
|
4Q5C
 
 | | TvNiR in complex with sulfite, middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-09-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be Published
|
|
6RHF
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain C85A variant (CagFbFP) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
6RHG
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
2K59
 
 | |
