1R55
 
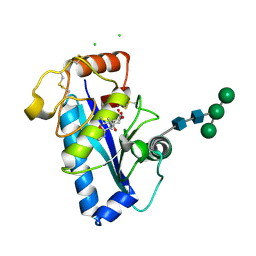 | | Crystal structure of the catalytic domain of human ADAM 33 | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reichert, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S, Le, H.V, Madison, V. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
2JM8
 
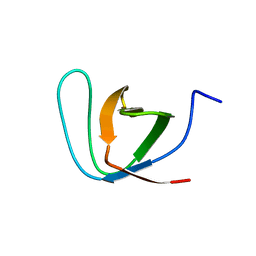 | | R21A Spc-SH3 free | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
1WT7
 
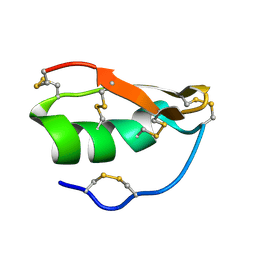 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
2JMA
 
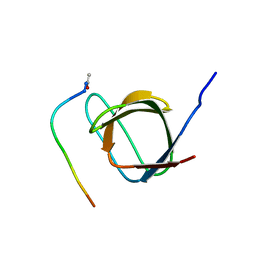 | | R21A Spc-SH3:P41 complex | | Descriptor: | P41 peptide, Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2JM9
 
 | | R21A Spc-SH3 bound | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
1R8G
 
 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1YNK
 
 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeteners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-[((R)-{[4-(AMINOMETHYL)PHENYL]AMINO}{[(1R)-1-PHENYLETHYL]AMINO}METHYL)AMINO]ETHANE-1,1-DIOL, Ig gamma heavy chain, immunoglobulin kappa light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
2IX6
 
 | | SHORT CHAIN SPECIFIC ACYL-COA OXIDASE FROM ARABIDOPSIS THALIANA, ACX4 | | Descriptor: | ACYL-COENZYME A OXIDASE 4, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mackenzie, J, Pedersen, L, Arent, S, Henriksen, A. | | Deposit date: | 2006-07-07 | | Release date: | 2006-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Controlling Electron Transfer in Acyl-Coa Oxidases and Dehydrogenases: A Structural View.
J.Biol.Chem., 281, 2006
|
|
1MWW
 
 | | THE STRUCTURE OF THE HYPOTHETICAL PROTEIN HI1388.1 FROM HAEMOPHILUS INFLUENZAE REVEALS A TAUTOMERASE/MIF FOLD | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, HYPOTHETICAL PROTEIN HI1388.1 | | Authors: | Lehmann, C, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the Hypothetical Protein HI1388.1 from Haemophilus influenzae
To be Published
|
|
2F2V
 
 | | alpha-spectrin SH3 domain A56G mutant | | Descriptor: | FORMIC ACID, Spectrin alpha chain, brain | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Casares, S, Lopez-Mayorga, O, Vega, C. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cooperative propagation of local stability changes from low-stability and high-stability regions in a SH3 domain
Proteins, 67, 2007
|
|
1HXD
 
 | | CRYSTAL STRUCTURE OF E. COLI BIOTIN REPRESSOR WITH BOUND BIOTIN | | Descriptor: | BIOTIN, BIRA BIFUNCTIONAL PROTEIN | | Authors: | Kwon, K, Streaker, E.D, Ruparelia, S, Beckett, D. | | Deposit date: | 2001-01-12 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1X6J
 
 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1X6I
 
 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
1RW1
 
 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1LCX
 
 | | NMR structure of HIV-1 gp41 659-671 13mer peptide | | Descriptor: | GP41 | | Authors: | Biron, Z, Khare, S, Samson, A.O, Hayek, Y, Naider, F, Anglister, J. | | Deposit date: | 2002-04-07 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Monomeric 3(10)-Helix Is Formed in Water by a 13-Residue Peptide
Representing the Neutralizing Determinant of HIV-1 on gp41.
Biochemistry, 41, 2002
|
|
