7ESL
 
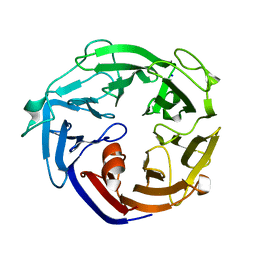 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, N247A N-glycan free form | | Descriptor: | L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
6KPM
 
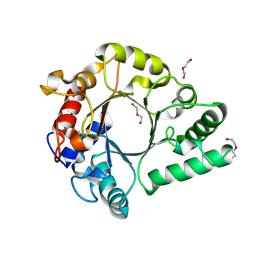 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPL
 
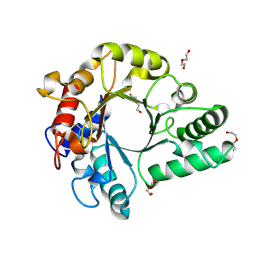 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
3WVL
 
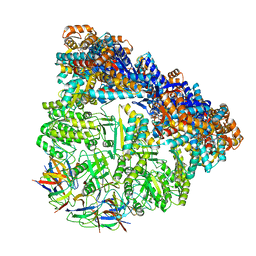 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
5AYI
 
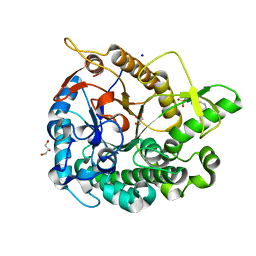 | | Crystal structure of GH1 Beta-glucosidase TD2F2 N223Q mutant | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-GLUCOSIDASE, GLYCEROL, ... | | Authors: | Jo, T, Manninen, J.A, Matsuzawa, T, Uchiyama, T, Yaoi, K, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-08-21 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2
Febs J., 283, 2016
|
|
5AYB
 
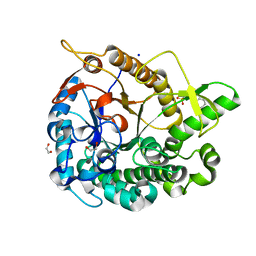 | | Crystal structure of GH1 Beta-Glucosidase TD2F2 N223G mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-GLUCOSIDASE, ... | | Authors: | Jo, T, Manninen, J.A, Matsuzawa, T, Uchiyama, T, Yaoi, K, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-08-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2
Febs J., 283, 2016
|
|
6LCE
 
 | |
6A0L
 
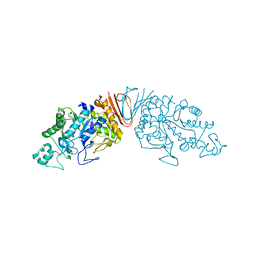 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with maltose | | Descriptor: | Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
5ZXG
 
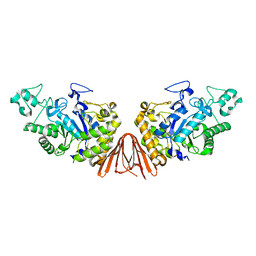 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, ligand-free form | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-05-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
6A0J
 
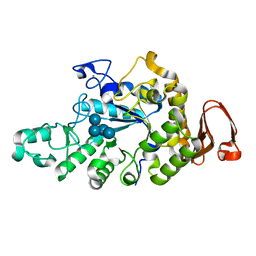 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with Cyclic alpha-maltosyl-(1-->6)-maltose | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
6A0K
 
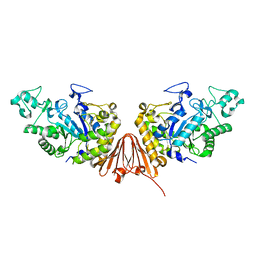 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with panose | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
5B0S
 
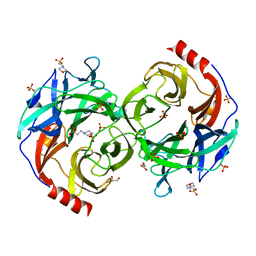 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannotriose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0R
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannobiose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0P
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - glycerol complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5B0Q
 
 | | beta-1,2-Mannobiose phosphorylase from Listeria innocua - mannose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lin0857 protein, SULFATE ION, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
4ZLG
 
 | | Cellobionic acid phosphorylase - gluconic acid complex | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLF
 
 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
6A94
 
 | | Crystal structure of 5-HT2AR in complex with zotepine | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(3-chloranylbenzo[b][1]benzothiepin-5-yl)oxy-N,N-dimethyl-ethanamine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6A93
 
 | | Crystal structure of 5-HT2AR in complex with risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3VYG
 
 | | Crystal structure of Thiocyanate hydrolase mutant R136W | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
4ZLE
 
 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5ZNM
 
 | | Colicin D Central Domain and C-terminal tRNase domain | | Descriptor: | Colicin-D, GLYCEROL, SULFATE ION | | Authors: | Chang, J.W, Sato, Y, Ogawa, T, Arakawa, T, Fukai, S, Fushinobu, S, Masaki, H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell
J. Biochem., 164, 2018
|
|
4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5B47
 
 | | 2-Oxoacid:Ferredoxin Oxidoreductase 2 from Sulfolobus tokodai - pyruvate complex | | Descriptor: | 2-oxoacid--ferredoxin oxidoreductase alpha subunit, 2-oxoacid--ferredoxin oxidoreductase beta subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Yan, Z, Maruyama, A, Arakawa, T, Fushinobu, S, Wakagi, T. | | Deposit date: | 2016-04-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of archaeal 2-oxoacid:ferredoxin oxidoreductases from Sulfolobus tokodaii
Sci Rep, 6, 2016
|
|
