2RSX
 
 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
1WZ7
 
 | | Crystal structure of enhancer of rudimentary homologue (ERH) | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Arai, R, Kukimoto-Niino, M, Uda-Tochio, H, Morita, S, Uchikubo-Kamo, T, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-26 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an enhancer of rudimentary homolog (ERH) at 2.1 Angstroms resolution.
Protein Sci., 14, 2005
|
|
1X42
 
 | | Crystal structure of a haloacid dehalogenase family protein (PH0459) from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PH0459 | | Authors: | Arai, R, Kukimoto-Niino, M, Sugahara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable haloacid dehalogenase PH0459 from Pyrococcus horikoshii OT3
Protein Sci., 15, 2006
|
|
2CZK
 
 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (trigonal form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Hanawa-Suetsugu, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2CZI
 
 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) with calcium and phosphate ions | | Descriptor: | CALCIUM ION, Inositol monophosphatase 2, PHOSPHATE ION | | Authors: | Arai, R, Ito, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2DDK
 
 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (orthorhombic form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Bessho, Y, Ohba, H, Ohnishi, T, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-30 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
3A3U
 
 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, L(+)-TARTARIC ACID, Menaquinone biosynthetic enzyme, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Uchikubo-Kamo, T, Katsura, K, Nishimoto, M, Toyama, M, Terada, T, Kuramitsu, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-06-20 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
2CZ5
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3
To be Published
|
|
2CWE
 
 | | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical transcription regulator protein, PH1932 | | Authors: | Arai, R, Kishishita, S, Kukimoto-Niino, M, Wang, H, Sugawara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3
To be Published
|
|
2CZL
 
 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 (Cys11 modified with beta-mercaptoethanol) | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Kamo-Uchikubo, T, Nishimoto, M, Toyama, M, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
2CZ4
 
 | | Crystal structure of a putative PII-like signaling protein (TTHA0516) from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, hypothetical protein TTHA0516 | | Authors: | Arai, R, Fusatomi, E, Kukimoto-Niino, M, Kawaguchi, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of a putative PII-like signaling protein (TTHA0516) from Thermus thermophilus HB8
To be Published
|
|
2CZE
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Arai, R, Ito, K, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP
To be Published
|
|
2CZD
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution
To be Published
|
|
3VJF
 
 | | Crystal structure of de novo 4-helix bundle protein WA20 | | Descriptor: | POTASSIUM ION, WA20 | | Authors: | Arai, R, Kimura, A, Kobayashi, N, Matsuo, K, Sato, T, Wang, A.F, Platt, J.M, Bradley, L.H, Hecht, M.H. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20
J.Phys.Chem.B, 116, 2012
|
|
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
6KXM
 
 | |
8H7E
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) at 2.0 angstrom resolution | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H7D
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H7C
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) - Pt derivative | | Descriptor: | ACETATE ION, CHLORIDE ION, De novo ferric enterobactin esterase Syn-F4, ... | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7EQ9
 
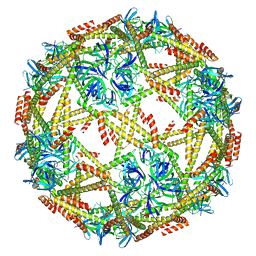 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | Descriptor: | TIP60 | | Authors: | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
7CYW
 
 | |
4YSH
 
 | |
6KOS
 
 | |
6KST
 
 | |
6KXN
 
 | | Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
