3SS9
 
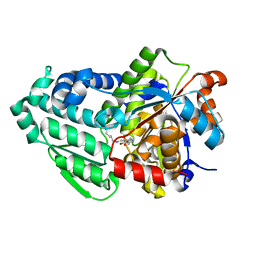 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
7ZCS
 
 | | Nitrite-bound MSOX movie series dataset 65 (52 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - water ligand (final) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCP
 
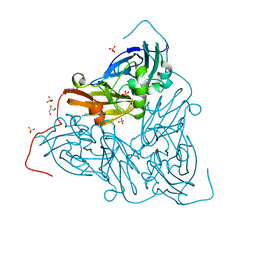 | | Nitrite-bound MSOX movie series dataset 17 (13.6 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitric oxide (NO) intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCO
 
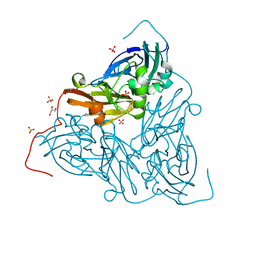 | | Nitrite-bound MSOX movie series dataset 8 (6.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitrite/NO intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCQ
 
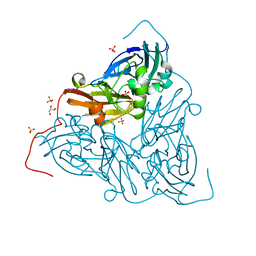 | | Nitrite-bound MSOX movie series dataset 25 (20 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - NO/water intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCR
 
 | | Nitrite-bound MSOX movie series dataset 38 (30.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - water ligand | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCN
 
 | | Nitrite-bound MSOX movie series dataset 1 (0.8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitrite (start) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QQ5
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric oxide reductase subunit B, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
2C9V
 
 | | Atomic resolution structure of Cu-Zn Human Superoxide dismutase | | Descriptor: | COPPER (II) ION, SODIUM ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2CNU
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with aspartic acid | | Descriptor: | ASPARTIC ACID, PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
2CNQ
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
2CNV
 
 | | SAICAR-synthase from Saccharomyces cerevisiae complexed SAICAR | | Descriptor: | ASPARTIC ACID, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID, PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, V.R. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Diffraction Study of the Complex of the Enzyme Saicar Synthase with the Reaction Product
Crystallogr.Rep.(Transl. Kristallografiya), 51, 2006
|
|
2C9U
 
 | | 1.24 Angstroms resolution structure of as-isolated Cu-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2C9S
 
 | | 1.24 Angstroms resolution structure of Zn-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
6ZAV
 
 | | NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.19 A resolution (unrestrained, full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAW
 
 | | Damage-free NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAR
 
 | | As-isolated copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.1 A resolution (unrestrained, full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6XVF
 
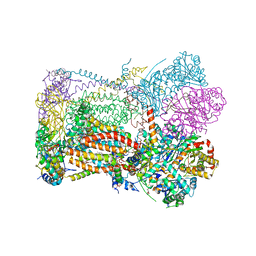 | | Crystal structure of bovine cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Tetrahydroquinolone Eliminates Apicomplexan Parasites.
Front Cell Infect Microbiol, 10, 2020
|
|
6ZAX
 
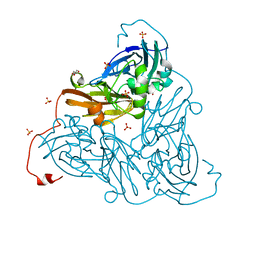 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at low dose (0.5 MGy) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAU
 
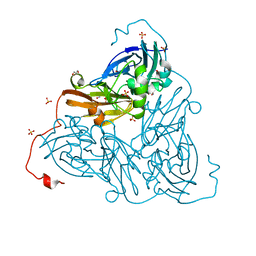 | | Damage-free nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAT
 
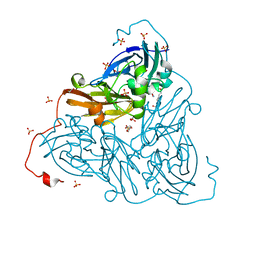 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.0 A resolution (unrestrained full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAS
 
 | | Damage-free as-isolated copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6QQ6
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
4CSP
 
 | | Structure of the F306C mutant of nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Leferink, N.G.H, Antonyuk, S.V, Houwman, J.A, Scrutton, N.S, REady, R, Hasnain, S.S. | | Deposit date: | 2014-03-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of Residues Remote from the Catalytic Centre on Enzyme Catalysis of Copper Nitrite Reductase.
Nat.Commun., 5, 2014
|
|
5NPL
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria, Yb-derivative at 2.8 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Similar to tr|Q8YYT1|Q8YYT1, YTTERBIUM (III) ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-30 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
