8TCA
 
 | | Structure of a mouse IgG antibody antigen-binding fragment (Fab) targeting N6-methyladenosine (m6A), an RNA modification, no ligand | | Descriptor: | 1,2-ETHANEDIOL, m6A binding IgG Fab, heavy chain, ... | | Authors: | Angelo, M.C, Aoki, S.T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of a mouse IgG antibody antigen-binding fragment (Fab) targeting N6-methyladenosine (m6A), an RNA modification
To Be Published
|
|
2GJ5
 
 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
4G2C
 
 | | DyP2 from Amycolatopsis sp. ATCC 39116 | | Descriptor: | ACETATE ION, DyP2, MANGANESE (II) ION, ... | | Authors: | Brown, M.E, Barros, T, Chang, M.C.Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium.
Acs Chem.Biol., 7, 2012
|
|
4GGO
 
 | |
1CC3
 
 | | PURPLE CUA CENTER | | Descriptor: | COPPER (II) ION, PROTEIN (CUA AZURIN) | | Authors: | Robinson, H, Ang, M.C, Gao, Y.-G, Hay, M.T, Lu, Y, Wang, A.H.-J. | | Deposit date: | 1999-03-03 | | Release date: | 1999-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of electron transfer modulation in the purple CuA center.
Biochemistry, 38, 1999
|
|
7TWA
 
 | | Crystal structure of apo BesC from Streptomyces cattleya | | Descriptor: | 1,3-BUTANEDIOL, 4-chloro-allylglycine synthase, ACETATE ION, ... | | Authors: | Neugebauer, M.E, McBride, M.J, Boal, A.K, Chang, M.C.Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate-Triggered mu-Peroxodiiron(III) Intermediate in the 4-Chloro-l-Lysine-Fragmenting Heme-Oxygenase-like Diiron Oxidase (HDO) BesC: Substrate Dissociation from, and C4 Targeting by, the Intermediate.
Biochemistry, 61, 2022
|
|
4GGP
 
 | |
3P3I
 
 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate and CoA | | Descriptor: | COENZYME A, Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2R
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate | | Descriptor: | Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2S
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in an open conformation | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P3F
 
 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2Q
 
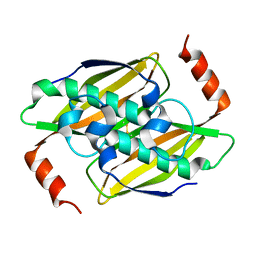 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase, FlK | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
5BUN
 
 | | Crystal structure of an antigenic outer membrane protein ST50 from Salmonella Typhi | | Descriptor: | Outer membrane protein, octyl beta-D-glucopyranoside | | Authors: | Yoshimura, M, Chuankhayan, P, Lin, C.C, Chen, N.C, Yang, M.C, Fun, H.K. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of an antigenic outer-membrane protein from Salmonella Typhi suggests a potential antigenic loop and an efflux mechanism.
Sci Rep, 5, 2015
|
|
7N09
 
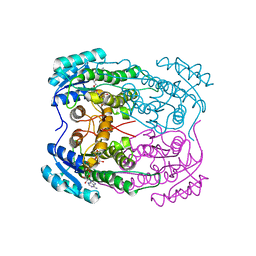 | |
7U6J
 
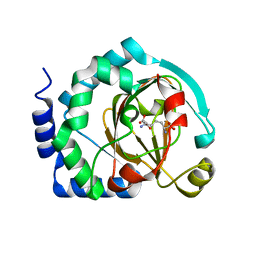 | | HalB with lysine and succinate | | Descriptor: | Halogenase B, LYSINE, SUCCINIC ACID | | Authors: | Sumida, K.H, Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7U6H
 
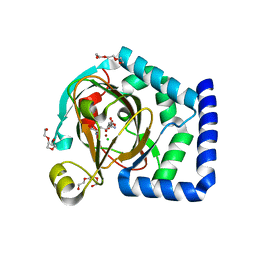 | | HalD with ornithine and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Swenson, C.V, Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6BJA
 
 | |
7JSD
 
 | | Hydroxylase homolog of BesD with Fe(II), alpha-ketoglutarate, and lysine | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, LYSINE, ... | | Authors: | Kissman, E.N, Neugebauer, M.E, Chang, M.C.Y. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction pathway engineering converts a radical hydroxylase into a halogenase.
Nat.Chem.Biol., 18, 2022
|
|
7U6I
 
 | | HalB with glycine and succinate | | Descriptor: | GLYCEROL, GLYCINE, Halogenase B, ... | | Authors: | Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6BJ9
 
 | |
6BJB
 
 | |
6NIE
 
 | | BesD with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, BesD, lysine halogenase, ... | | Authors: | Neugebauer, M.E, Chang, M.C.Y. | | Deposit date: | 2018-12-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A family of radical halogenases for the engineering of amino-acid-based products.
Nat.Chem.Biol., 15, 2019
|
|
