1DKC
 
 | | SOLUTION STRUCTURE OF PAFP-S, AN ANTIFUNGAL PEPTIDE FROM THE SEEDS OF PHYTOLACCA AMERICANA | | Descriptor: | ANTIFUNGAL PEPTIDE | | Authors: | Wang, D.C, Gao, G.H, Shao, F, Dai, J.X, Wang, J.F. | | Deposit date: | 1999-12-07 | | Release date: | 2000-12-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PAFP-S: a new knottin-type antifungal peptide from the seeds of Phytolacca americana
Biochemistry, 40, 2001
|
|
7KJQ
 
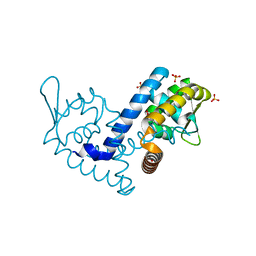 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Picloram | | Descriptor: | Picloram, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KH3
 
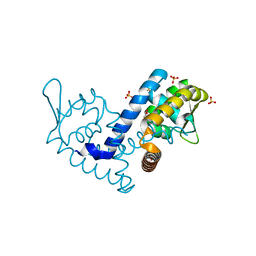 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 propionic acid | | Descriptor: | INDOLYLPROPIONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KK0
 
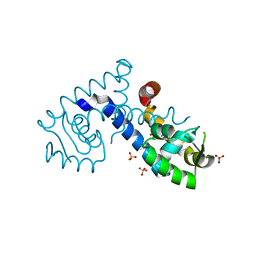 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Catechol | | Descriptor: | CATECHOL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KUA
 
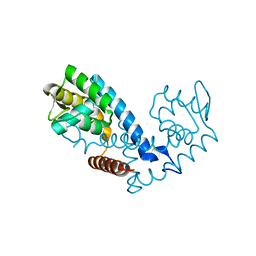 | | Crystal structure of the MarR family transcriptional regulator from Pseudomonas putida bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Transcriptional regulator, MarR family | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KKC
 
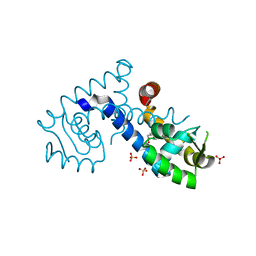 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 5-Hydroxyindoleacetic acid | | Descriptor: | (5-hydroxy-1H-indol-3-yl)acetic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KRH
 
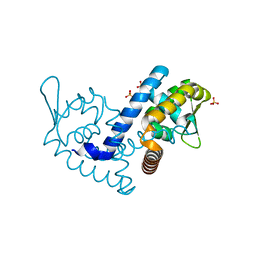 | |
7KJL
 
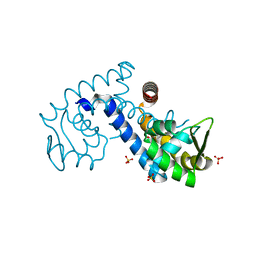 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KKI
 
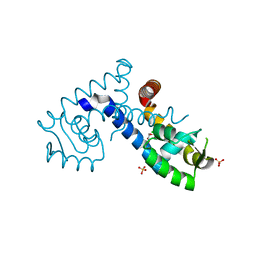 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 2,4-Dichlorophenoxyacetic acid | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KIG
 
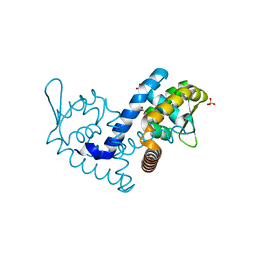 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole-3-butyric acid | | Descriptor: | 3-INDOLEBUTYRIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KYM
 
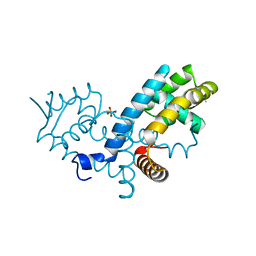 | |
2H7B
 
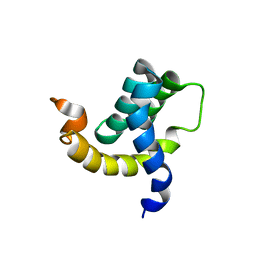 | | Solution structure of the eTAFH domain from the human leukemia-associated fusion protein AML1-ETO | | Descriptor: | Core-binding factor, ML1-ETO | | Authors: | Plevin, M.J, Zhang, J, Guo, C, Roeder, R.G, Ikura, M. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7L19
 
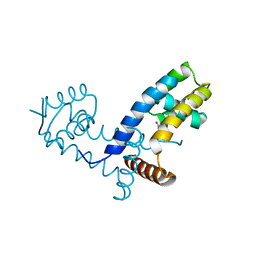 | | Crystal structure of the MarR family transcriptional regulator from Enterobacter soli strain LF7 bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family transcriptional regulator, NICKEL (II) ION | | Authors: | Lietzan, A.D, Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
2HEI
 
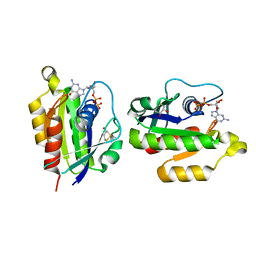 | | Crystal structure of human RAB5B in complex with GDP | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5B | | Authors: | Hong, B, Shen, L, Wang, J, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human RAB5B in complex with GDP
To be Published
|
|
1NY8
 
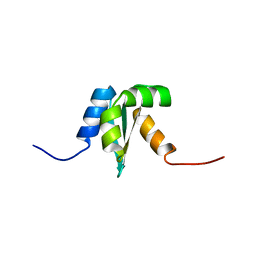 | | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115 | | Descriptor: | Protein yrbA | | Authors: | Swapna, G.V.T, Huang, J.Y, Acton, T.B, Shastry, R, Chiang, Y.-W, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115
To be Published
|
|
3C7N
 
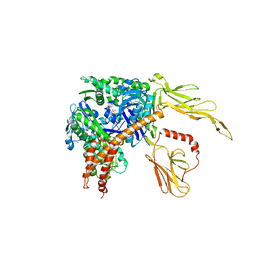 | | Structure of the Hsp110:Hsc70 Nucleotide Exchange Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Schuermann, J.P, Jiang, J, Hart, P.J, Sousa, R. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | Structure of the Hsp110:Hsc70 nucleotide exchange machine
Mol.Cell, 31, 2008
|
|
3CCN
 
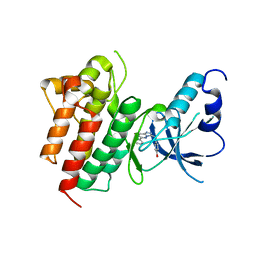 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | Descriptor: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | Authors: | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
2KQB
 
 | | First PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KLI
 
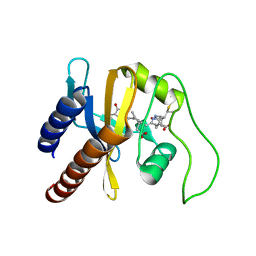 | | Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Zhang, J, Rivera, M, Vierstra, R.D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-03 | | Release date: | 2009-11-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form
Nature, 463, 2010
|
|
2KMI
 
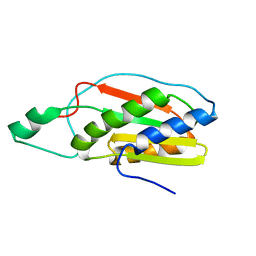 | |
3Q2J
 
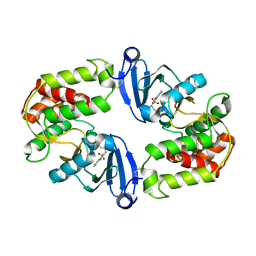 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Protein Kinase Inhibitor CKI-7 Complex | | Descriptor: | Aminoglycoside 3'-phosphotransferase, CALCIUM ION, N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE | | Authors: | Fong, D.H, Xiong, B, Hwang, J, Berghuis, A.M. | | Deposit date: | 2010-12-20 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1501 Å) | | Cite: | Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor.
Plos One, 6, 2011
|
|
7L1I
 
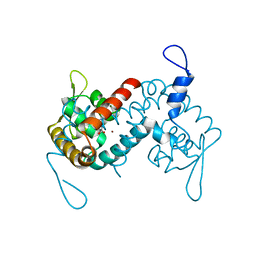 | | Crystal structure of the MarR family transcriptional regulator from Acineotobacter baumannii bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family multidrug resistance pump transcriptional regulator, NICKEL (II) ION | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
1B03
 
 | |
7L83
 
 | | NMR solution structure of Nav1.5 DIV S3b-S4a paddle motif in DPC micelle | | Descriptor: | Sodium channel protein type 5 subunit alpha | | Authors: | Hussein, A.K, Bhuiyan, M.H, Arshava, B, Zhuang, J, Poget, S.F. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and analysis of isolated S3b-S4a motif of repeat IV of the human cardiac sodium channel
Biorxiv, 2021
|
|
2KQD
 
 | | First PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
