1C5S
 
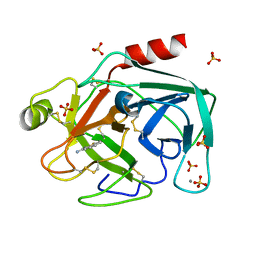 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, PROTEIN (TRYPSIN), ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5U
 
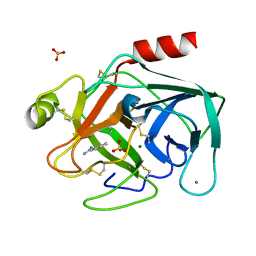 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PROTEIN (TRYPSIN), ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5R
 
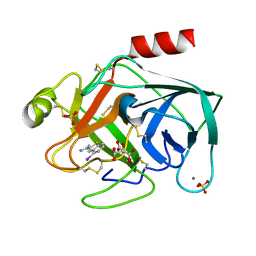 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
7JNH
 
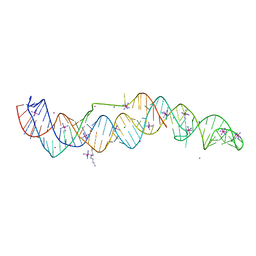 | | Crystal structure of a double-ENE RNA stability element in complex with a 28-mer poly(A) RNA | | Descriptor: | 28-mer poly(A) RNA, COBALT HEXAMMINE(III), Core double ENE RNA (Xtal construct) from Oryza sativa transposon,Core double ENE RNA (Xtal construct) from Oryza sativa transposon, ... | | Authors: | Torabi, S.F, Vaidya, A.T, Tycowski, K.T, DeGregorio, S.J, Wang, J, Shu, M.D, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction.
Science, 371, 2021
|
|
3I0Q
 
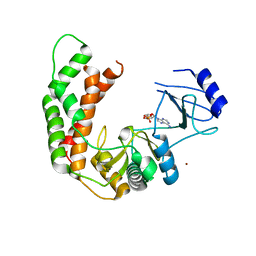 | | Crystal Structure of the AMP-bound complex of Spectinomycin Phosphotransferase, APH(9)-Ia | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICKEL (II) ION, Spectinomycin phosphotransferase | | Authors: | Berghuis, A.M, Fong, D.H, Lemke, C.T, Hwang, J.-Y, Xiong, B. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
6GGY
 
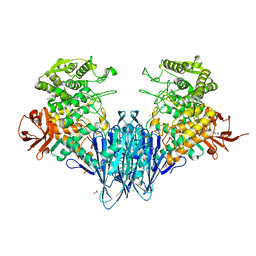 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with sulphate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Laminaribiose phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
3NWN
 
 | | Crystal structure of the human KIF9 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF9, ... | | Authors: | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human KIF9 motor domain in complex with ADP
To be Published
|
|
4OR9
 
 | | Crystal structure of human calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
1K8M
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
5IVW
 
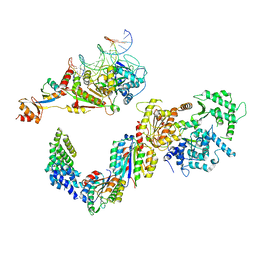 | | Human core TFIIH bound to DNA within the PIC | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, General transcription factor IIH subunit 4, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-21 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
3V6P
 
 | | Crystal structure of the DNA-binding domain of dHax3, a TAL effector | | Descriptor: | dHax3 | | Authors: | Deng, D, Yan, C.Y, Pan, X.J, Wang, J.W, Shi, Y.G, Yan, N. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Sequence-Specific Recognition of DNA by TAL Effectors
Science, 2012
|
|
3I1A
 
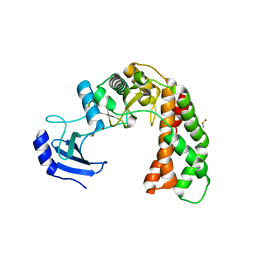 | | Crystal Structure of apo Spectinomycin Phosphotransferase, APH(9)-Ia | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, Spectinomycin phosphotransferase, ... | | Authors: | Berghuis, A.M, Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
1KUY
 
 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
3K6W
 
 | | Apo and ligand bound structures of ModA from the archaeon Methanosarcina acetivorans | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6X
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Molybdate-Bound Close Form with 2 Molecules in Asymmetric Unit Forming Beta Barrel | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7JW6
 
 | | Crystal structure of Drosophila Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3DJE
 
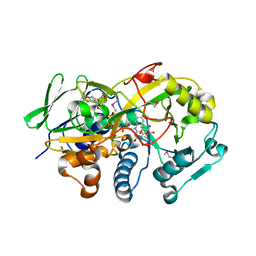 | | Crystal structure of the deglycating enzyme fructosamine oxidase from Aspergillus fumigatus (Amadoriase II) in complex with FSA | | Descriptor: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Collard, F, Zhang, J, Nemet, I, Qanungo, K.R, Monnier, V.M, Yee, V.C. | | Deposit date: | 2008-06-23 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the deglycating enzyme fructosamine oxidase (FAOX-II)
To be Published
|
|
8P1S
 
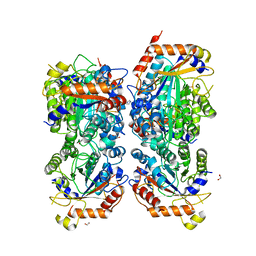 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) in complex with fucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819), ... | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
6GH3
 
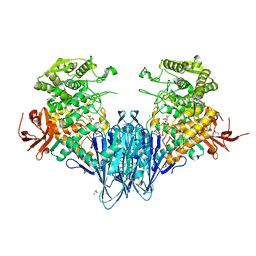 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-man-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
5T09
 
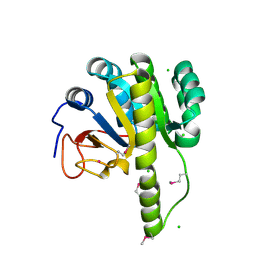 | | The structure of the type III effector HopBA1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Type III secretion system effector HopBA1 | | Authors: | Cherkis, K, Machius, M, Nishimura, M.T, Dangl, J.L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7JVD
 
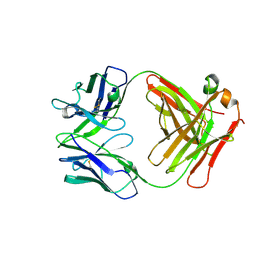 | | Fab of 5.6 monoclonal mouse IgG1 co-crystallized with the trisaccharide form of serotype 3 pneumococcal capsular polysaccharide | | Descriptor: | 5.6 Fab heavy chain, 5.6 Fab light chain, beta-D-glucopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose | | Authors: | Ozdilek, A, Huang, J, Paschall, A.V, Babb, R, Middleton, D.R, Duke, J.A, Pirofski, L, Mousa, J.J, Avci, F.Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Structural Model for the Ligand Binding of Pneumococcal Serotype 3 Capsular Polysaccharide-Specific Protective Antibodies.
Mbio, 12, 2021
|
|
5XVC
 
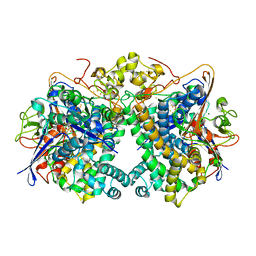 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
3JAY
 
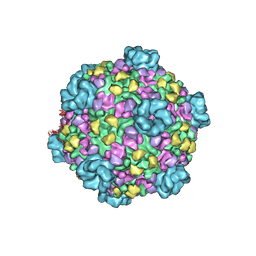 | | Atomic model of transcribing cytoplasmic polyhedrosis virus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capsid protein VP1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
3JB1
 
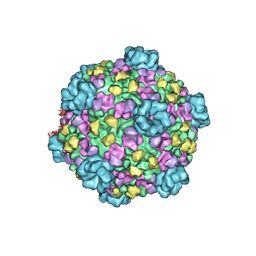 | | Atomic model of cytoplasmic polyhedrosis virus with SAM | | Descriptor: | Capsid protein VP1, S-ADENOSYLMETHIONINE, Structural protein VP3, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
4NCB
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 19-mer target DNA with Mg2+ | | Descriptor: | 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3', 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
