8HPG
 
 | |
4NXB
 
 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
5Z11
 
 | | Crystal Structure of Grass Carp CD8 alpha alpha Homodimers | | Descriptor: | CD8 alpha chain | | Authors: | Wang, J. | | Deposit date: | 2017-12-22 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Grass Carp CD8 alpha
alpha Homodimers Indicates a Dramatic Evolution of CD8 from Ectotherms to Endotherms.
To Be Published
|
|
5ZCJ
 
 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
8JJG
 
 | | Crystal structure of QW-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJI
 
 | | Crystal structure of QR-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK0
 
 | | Crystal structure of QL-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJH
 
 | | Crystal structure of QH-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJX
 
 | | Crystal structure of QS-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJF
 
 | | Crystal structure of QE-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJU
 
 | | Crystal structure of QD-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJZ
 
 | | Crystal structure of QQ-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK1
 
 | | Crystal structure of QA-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
6BJS
 
 | | CryoEM structure of E.coli his pause elongation complex without pause hairpin | | Descriptor: | DNA (32-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Landick, R, Darst, S.A. | | Deposit date: | 2017-11-06 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing.
Mol. Cell, 69, 2018
|
|
6C1I
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with T0070907 | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Fuhrmann, J, Brust, R, Kojetin, D.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A structural mechanism for directing corepressor-selective inverse agonism of PPAR gamma.
Nat Commun, 9, 2018
|
|
4L53
 
 | | Crystal Structure of (1R,4R)-4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)-3-chlorofuro[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}cyclohexan-1-ol bound to TAK1-TAB1 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, ... | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1: Optimization of kinase selectivity and pharmacokinetics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3U86
 
 | |
5SVD
 
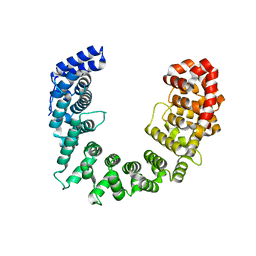 | |
6C6S
 
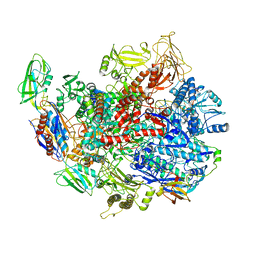 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
3PLS
 
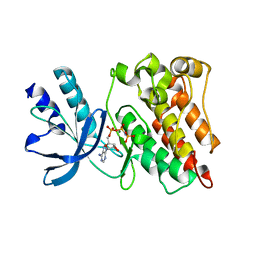 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
6K3E
 
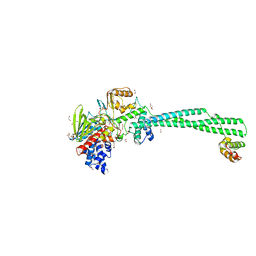 | | LSD1/Co-Rest structure with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PCPA derivative, ... | | Authors: | Wang, J. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | LSD1/Co-Rest structure with an inhibitor
To Be Published
|
|
8ILJ
 
 | |
4DNC
 
 | | Crystal structure of human MOF in complex with MSL1 | | Descriptor: | Histone acetyltransferase KAT8, Male-specific lethal 1 homolog, ZINC ION | | Authors: | Huang, J, Lei, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the regulation of MOF in the male-specific lethal complex and the non-specific lethal complex.
Cell Res., 22, 2012
|
|
6C6T
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
7VMC
 
 | | Crystal structure of EF-Tu/CdiA/CdiI | | Descriptor: | Contact-dependent inhibitor I, Elongation factor Tu, tRNA nuclease CdiA | | Authors: | Wang, J, Yashiro, Y, Tomita, K. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.413 Å) | | Cite: | Mechanistic insights into tRNA cleavage by a contact-dependent growth inhibitor protein and translation factors.
Nucleic Acids Res., 50, 2022
|
|
