3LYL
 
 | | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-27 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis
TO BE PUBLISHED
|
|
1OTB
 
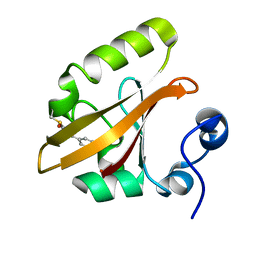 | | WILD TYPE PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OT6
 
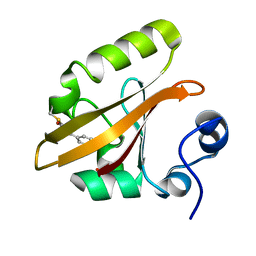 | |
1OTD
 
 | |
1OTE
 
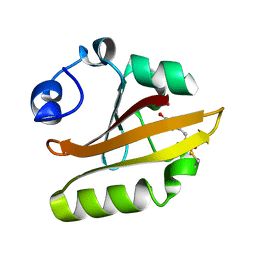 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, photoactive yellow protein, PYP | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTA
 
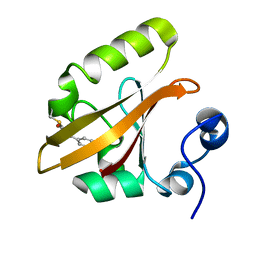 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OT9
 
 | | CRYOTRAPPED STATE IN WILD TYPE PHOTOACTIVE YELLOW PROTEIN, INDUCED WITH CONTINUOUS ILLUMINATION AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTI
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1YRX
 
 | | Structure of a novel photoreceptor: the BLUF domain of AppA from Rhodobacter sphaeroides | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-DODECYL-N,N-DIMETHYLGLYCINATE, hypothetical protein Rsph03001874 | | Authors: | Anderson, S, Dragnea, V, Masuda, S, Ybe, J, Moffat, K, Bauer, C. | | Deposit date: | 2005-02-05 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Novel Photoreceptor, the BLUF Domain of AppA from Rhodobacter sphaeroides
Biochemistry, 44, 2005
|
|
1S1Y
 
 | | Photoactivated chromophore conformation in Photoactive Yellow Protein (E46Q mutant) from 10 microseconds to 3 milliseconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Srajer, V, Pahl, R, Rajagopal, S, Schotte, F, Anfinrud, P, Wulff, M, Moffat, K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein
Structure, 12, 2004
|
|
1S1Z
 
 | | Photoactivated chromophore conformation in Photoactive Yellow Protein (E46Q mutant) from 10 to 500 nanoseconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive Yellow Protein | | Authors: | Anderson, S, Srajer, V, Pahl, R, Rajagopal, S, Schotte, F, Anfinrud, P, Wulff, M, Moffat, K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein
Structure, 12, 2004
|
|
3KUU
 
 | | Structure of the PurE Phosphoribosylaminoimidazole Carboxylase Catalytic Subunit from Yersinia pestis | | Descriptor: | Phosphoribosylaminoimidazole carboxylase catalytic subunit PurE, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure of the PurE Phosphoribosylaminoimidazole Carboxylase Catalytic Subunit from Yersinia pestis
To be Published
|
|
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
3IG4
 
 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
4EGU
 
 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
3GEU
 
 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3HZN
 
 | | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase
TO BE PUBLISHED
|
|
3LAC
 
 | | Crystal structure of Bacillus anthracis pyrrolidone-carboxylate peptidase, pcP | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Pyrrolidone-carboxylate peptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus anthracis pyrrolidone-carboxylate peptidase, pcP
To be Published
|
|
3L44
 
 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
3K1S
 
 | | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PTS system, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis
To be Published
|
|
3KOM
 
 | | Crystal structure of apo transketolase from Francisella tularensis | | Descriptor: | Transketolase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Gordon, E, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-13 | | Release date: | 2009-12-01 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of apo transketolase from Francisella tularensis
TO BE PUBLISHED
|
|
4O96
 
 | | 2.60 Angstrom resolution crystal structure of a protein kinase domain of type III effector NleH2 (ECs1814) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, type III effector protein kinase | | Authors: | Anderson, S.M, Halavaty, A.S, Wawrzak, Z, Kudritska, M, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type III Effector NleH2 from Escherichia coli O157:H7 str. Sakai Features an Atypical Protein Kinase Domain.
Biochemistry, 53, 2014
|
|
3LU2
 
 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
3KUX
 
 | | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis
To be Published
|
|
