7PB6
 
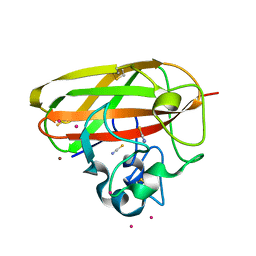 | |
4RW7
 
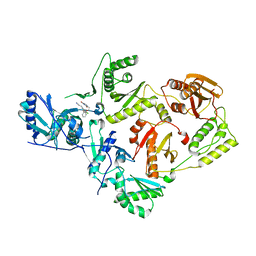 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) variant in complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ532), a non-nucleoside inhibitor | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-12-01 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.014 Å) | | Cite: | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
7M90
 
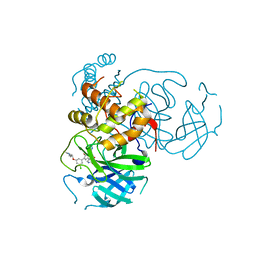 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8O
 
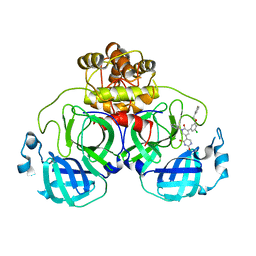 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
8B9V
 
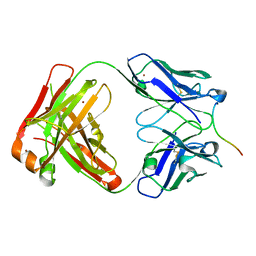 | | Crystal structure of Lu AF82422 in complex with alpha-synuclein 110-120 | | Descriptor: | Alpha-synuclein 110-120, Lu AF82422 Fab heavy chain, Lu AF82422 Fab light chain, ... | | Authors: | Bjerregaard-Andersen, K, Carr, K, Krogh, B.O, Kallunki, P, Tagmose, L, David, L. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Lu AF82422 in complex with alpha-synuclein 110-120
To Be Published
|
|
7M8Y
 
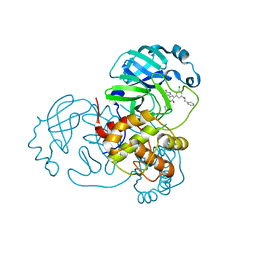 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 15 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M91
 
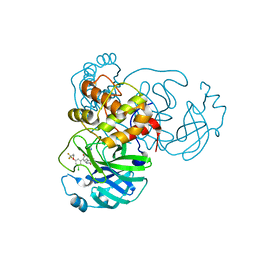 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 25 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8M
 
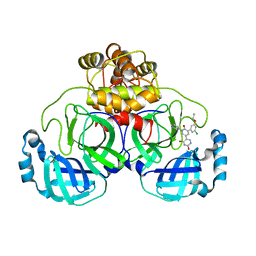 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 3C-like proteinase, 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8X
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Z
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3-hydroxy-3-methylbutoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8P
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 23 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8N
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
6EHG
 
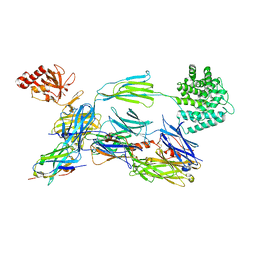 | | complement component C3b in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, K.R, Gadeberg, T.A.F, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement.
J. Biol. Chem., 293, 2018
|
|
1ACA
 
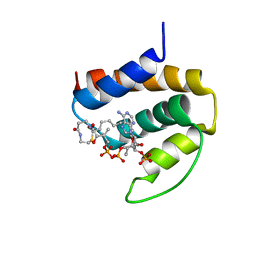 | | THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX BETWEEN ACYL-COENZYME A BINDING PROTEIN AND PALMITOYL-COENZYME A | | Descriptor: | ACYL-COENZYME A BINDING PROTEIN, COENZYME A, PALMITIC ACID | | Authors: | Kragelund, B.B, Andersen, K.V, Madsen, J.C, Knudsen, J, Poulsen, F.M. | | Deposit date: | 1992-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the complex between acyl-coenzyme A binding protein and palmitoyl-coenzyme A.
J.Mol.Biol., 230, 1993
|
|
5NNA
 
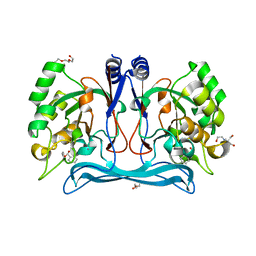 | | Isatin hydrolase A (IHA) from Labrenzia aggregata bound to benzyl benzoate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZOIC ACID PHENYLMETHYLESTER, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sommer, T, Bjerregaard-Andersen, K, Morth, J.P. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A fundamental catalytic difference between zinc and manganese dependent enzymes revealed in a bacterial isatin hydrolase.
Sci Rep, 8, 2018
|
|
5NMP
 
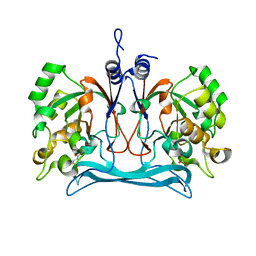 | |
1A3H
 
 | |
7AU7
 
 | | Crystal structure of Nod Factor Perception ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BAX
 
 | | Crystal structure of LYS11 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase | | Authors: | Laursen, M, Cheng, J, Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-12-16 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4RWI
 
 | |
7PB7
 
 | |
5NNB
 
 | |
5NML
 
 | | Nb36 Ser85Cys with Hg bound | | Descriptor: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6S2I
 
 | |
5JJ2
 
 | | Crystal structure of the central domain of human AKAP18 gamma/delta in complex with malonate | | Descriptor: | A-kinase anchor protein 7 isoform gamma, MALONATE ION | | Authors: | Bjerregaard-Andersen, K, Ostensen, E, Scott, J.D, Tasken, K, Morth, J.P. | | Deposit date: | 2016-04-22 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Malonate in the nucleotide-binding site traps human AKAP18 gamma / delta in a novel conformational state.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
