3NVS
 
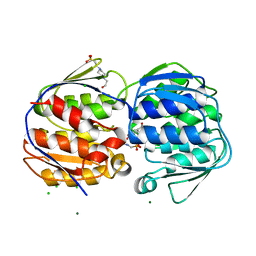 | | 1.02 Angstrom resolution crystal structure of 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae in complex with shikimate-3-phosphate (partially photolyzed) and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.021 Å) | | Cite: | 1.02 Angstrom Resolution Crystal Structure of 3-Phosphoshikimate 1-Carboxyvinyltransferase from Vibrio cholerae in complex with Shikimate-3-Phosphate (Partially Photolyzed) and Glyphosate
TO BE PUBLISHED
|
|
3NXK
 
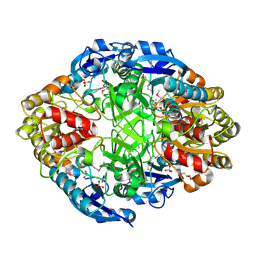 | | Crystal Structure of Probable Cytoplasmic L-asparaginase from Campylobacter jejuni | | Descriptor: | ACETIC ACID, Cytoplasmic L-asparaginase, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Probable Cytoplasmic L-asparaginase from Campylobacter jejuni
To be Published
|
|
3NTX
 
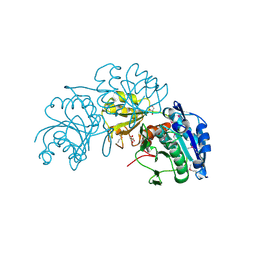 | | Crystal Structure of L-asparaginase I from Yersinia pestis | | Descriptor: | Cytoplasmic L-asparaginase I, GLYCEROL | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-05 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L-asparaginase I from Yersinia pestis
To be Published, 2010
|
|
4WZU
 
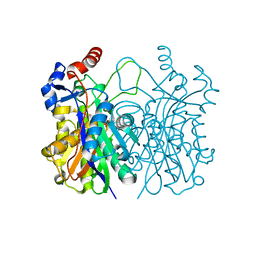 | | Crystal structure of beta-ketoacyl-(acyl carrier protein) synthase III-2 (FabH2) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, SODIUM ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Chordia, M.D, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
To Be Published
|
|
4X0O
 
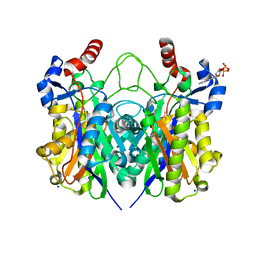 | | Beta-ketoacyl-(acyl carrier protein) synthase III-2 (FabH2) from Vibrio cholerae soaked with Acetyl-CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Chordia, M.D, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
4X3Z
 
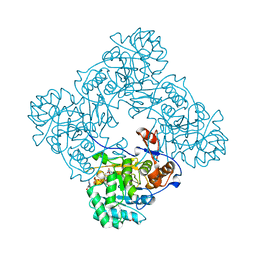 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
4XCW
 
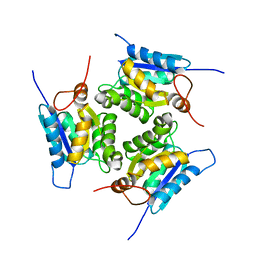 | |
4MHB
 
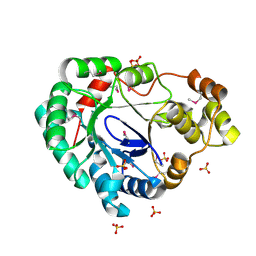 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
4O96
 
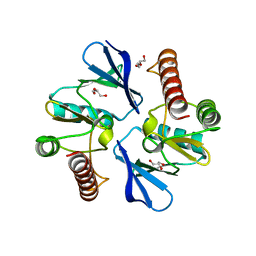 | | 2.60 Angstrom resolution crystal structure of a protein kinase domain of type III effector NleH2 (ECs1814) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, type III effector protein kinase | | Authors: | Anderson, S.M, Halavaty, A.S, Wawrzak, Z, Kudritska, M, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type III Effector NleH2 from Escherichia coli O157:H7 str. Sakai Features an Atypical Protein Kinase Domain.
Biochemistry, 53, 2014
|
|
4ZXU
 
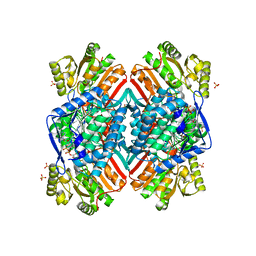 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
3OSU
 
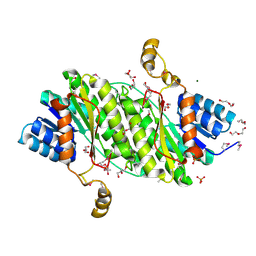 | | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Edwards, A, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-09 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus
To be Published
|
|
3R3R
 
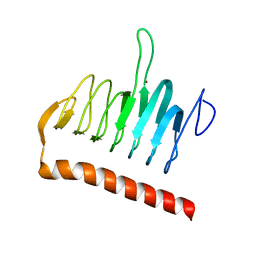 | | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica | | Descriptor: | ZINC ION, ferripyochelin binding protein | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica
TO BE PUBLISHED
|
|
5T1Q
 
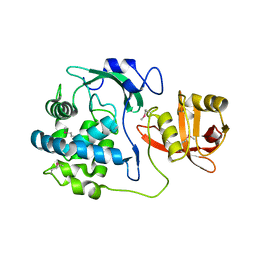 | | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus. | | Descriptor: | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979, SODIUM ION, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
To Be Published
|
|
5SWU
 
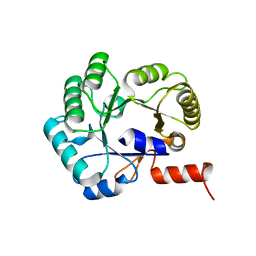 | |
5T1P
 
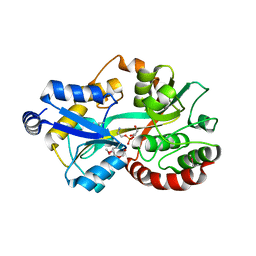 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
5SWV
 
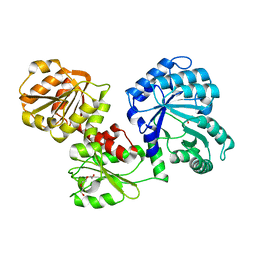 | |
5SV5
 
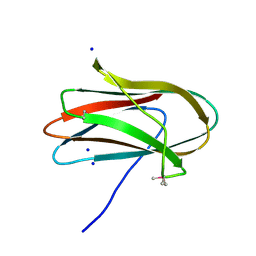 | | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis. | | Descriptor: | Microbial collagenase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis.
To Be Published
|
|
4EGU
 
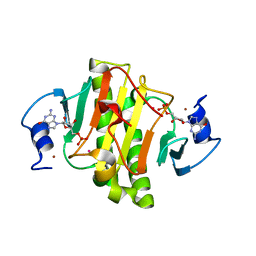 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
3Q7H
 
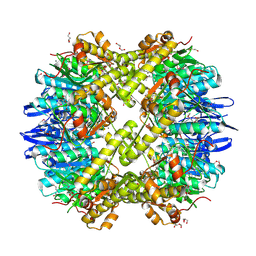 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
3R3S
 
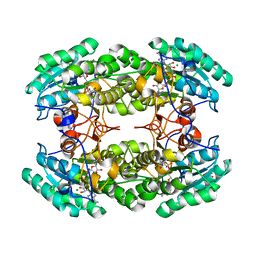 | | Structure of the YghA Oxidoreductase from Salmonella enterica | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the YghA Oxidoreductase from Salmonella enterica
TO BE PUBLISHED
|
|
5T1V
 
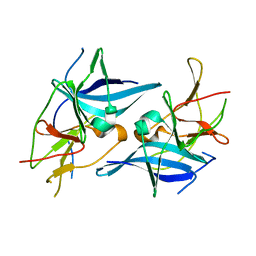 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | Descriptor: | NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
5T79
 
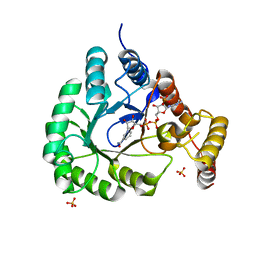 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | Descriptor: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
3R8Y
 
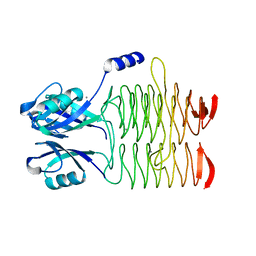 | | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, CALCIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase
TO BE PUBLISHED
|
|
5T8K
 
 | | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD.
To Be Published
|
|
5TJ9
 
 | | 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD | | Descriptor: | (1R,2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol, Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD
To Be Published
|
|
